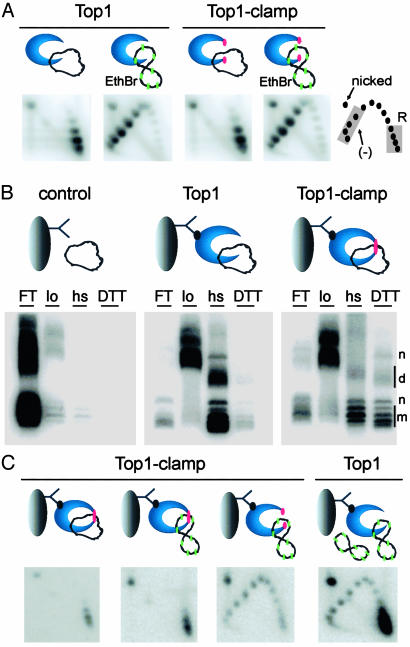
Fig. 5.
DNA rotation is inhibited in locked Top1-Clamp–DNA complexes. (A) Top1 and Top1-Clamp were incubated with relaxed plasmid DNA in G-100 buffer. EtdBr (green) was added to induce positive supercoils. After 30 min at 37°C, reactions were terminated and plasmid DNA topoisomers were resolved in 2D gels. Relaxed (R) and negatively supercoiled (–) DNAs are indicated. (B) Top1 and Top1-Clamp, bound to cellulose beads by the M2 antibody (12), were incubated with relaxed plasmid DNA, followed by diamide. DNA in the flow-through (FT), low-salt (lo), hs, and DTT/1 M KCl (DTT) washes were resolved in an agarose gel and visualized with Southern blotting. Monomer (m), nicked (n), and dimer (d) DNAs are indicated. Anomalies in DNA migration (FT and lo) were an artifact of protein contamination. (C) Bead-bound Top1 and Top1-Clamp hs samples were equilibrated with G-100 buffer in the presence or absence of diamide. Locked Top1-Clamp (single red dot) was treated with EtdBr (green) to induce positive supercoils. The sample lacking diamide was treated with EtdBr plus DTT to unlock the clamp (double red dots). Relaxed plasmid DNA and EtdBr were added to a similar Top1 sample. After 30 min at 37°C, reactions were terminated with DTT/1 M KCl/SDS, and DNA topoisomers were resolved in 2D gels.