Figure 2.
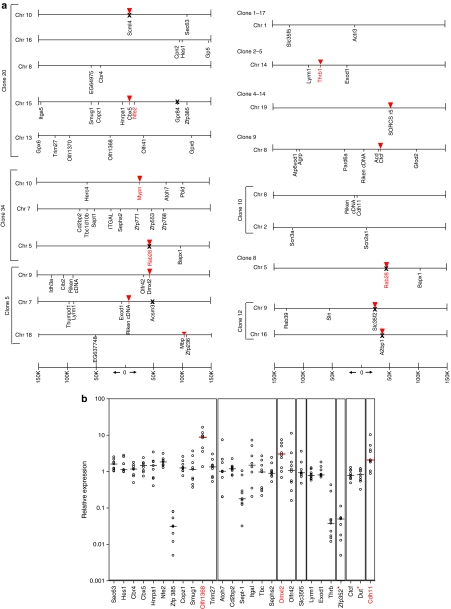
Insertional site analysis and expression of surrounding genes in clones derived from LCR β-GFP LV. (a) Schematic representation of the genomic annotation scale ±150 kb around the viral integration site is shown at the bottom. Inverted triangles (red) illustrate the integration site in the clones near genes. In gene bare regions, the insertion site is at 0, according to the scale beneath the clones. The recovered genes surrounding the integration site is marked in black. Genes highlighted in red indicate genes implicated in cancer. ‘X' represents genes analyzed for expression by quantitative reverse-transcriptase PCR but were not expressed. (b) Quantitative RT-PCR analysis on the surrounding gene expression in an immortalized clone with a specific insertion site. Expression of genes flanking the provirus was determined using real-time RT-PCR using the TaqMan low-density custom arrays. Expression was first normalized to the level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA expression. Alteration in the surrounding gene expression in an immortalized clone with a specific insertion site was calculated by comparing it against other immortalized clones containing vector insertions at other locations. The SDS RQ Manager 1.2 Software (Applied Biosystems, Foster City, CA) was used to calculate relative expression levels. This type of analysis was done to insure that the effect of gene expression was specific to the unique insertion site and not due to chance. This real-time RT-PCR data from individual genes thus analyzed was plotted as relative gene expression (log10 variations) and compared against other immortalized, unrelated clones containing vector insertions at other locations (shown as empty circles). The horizontal black line represents the median level of expression of the immortalized clone compared against expression from other clones with unrelated insertion sites. The horizontal red line and genes in red indicate median level of expression from significantly upregulated genes. y axis represents expression levels as relative mRNA quantity after normalization for the level of glyceraldehyde-3-phosphate dehydrogenase, and plotted as log10 variations from the median level in all analyzed clones. The genes marked with * are located at 435 kb (Zfp352) and 206 kb (Dut) from insertion site.