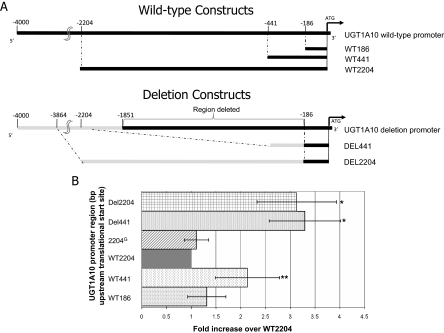
Fig. 3.
Role of the UGT1A10 promoter deletion in regulating heterologous gene expression. A, schematic representation of the PCR amplification strategy used for the UGT1A10 promoter luciferase assays. Top constructs illustrate the PCR-amplified fragments containing 186, 441, or 2204 bp of wild-type (WT) UGT1A10 promoter. The bottom constructs illustrate the PCR-amplified fragments containing either 441 or 2204 bp of deletion (DEL) variant UGT1A10 promoter. The gray lines indicate the promoter region located directly upstream of the UGT1A10 deletion; UGT1A10 deletion-containing luciferase vectors were constructed so that the UGT1A10 deletion and wild-type promoters were the same size. B, UGT1A10 wild-type, UGT1A10–2204G (containing 2204 bp of adjacent wild-type UGT1A10 promoter sequences except with a C>G SNP located at bp −1271 upstream of the UGT1A10 translation start site), and UGT1A10 deletion promoter regulation of luciferase activity in Caco-2 cells. Data are presented relative to the WT2204-overexpressing cell line. Bars indicate S.D. *, p = 0.009; **, p = 0.048, a significant increase in luciferase activity was observed compared with the WT2204-overexpressing cell line.