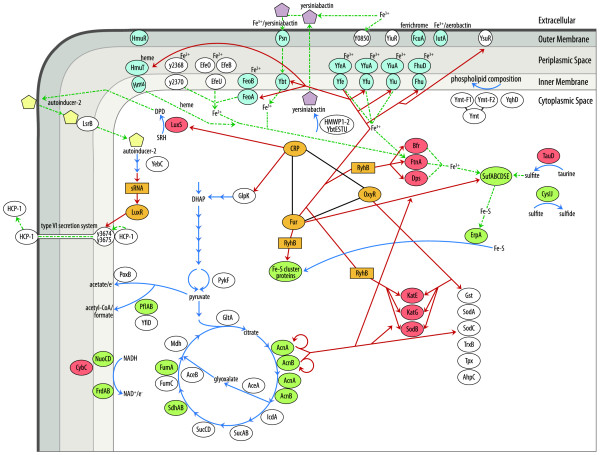
Figure 5.
Iron homeostasis in Y. pestis. The center of the schematic depicts a network of regulators (orange color), known or potentially involved in iron homeostasis. Details are provided in the text. CRP (carbon metabolism); OxyR (oxidative stress); Fur and small RNAs like RyhB (iron homeostasis). Red lines/arrows show which genes (or mRNAs) are controlled by these regulators. Additional arrows symbolize enzymatic reactions (blue line) or small molecule transport processes (dotted green line). The lower/left side of the schematic depicts components of the energy metabolism. It includes glycolytic steps from dihydroxyacetone phosphate (DHAP) to pyruvate, the TCA/glyoxalate bypass cycle and on the left side alternative pyruvate metabolism branches generating acetate or acetyl-CoA. Subunits of electron transport systems (NuoCD, FrdAB and CybC) are also displayed. The top/left side of the schematic pertains to quorum sensing. LuxS converts S-ribosylhomocysteine (SRH) to 4,5-dihydroxy-2,3-pentanedione (DPD) which is a precursor of autoinducer-2 (yellow pentagon). In E. coli, the autoinducer-2 is exported and imported via periplasmic LsrB into different cells followed by activation of LuxR via small RNA regulators. The precise functional role of YebC in quorum sensing is not known. LuxR influences the expression of virulence factors in pathogenic E. coli strains. The role of LuxR in the regulation of the type VI secretion system is speculative, but both iron starvation [73] and the T6SS [74] have been linked to quorum sensing in other organisms. In the upper part of the schematic, iron transporter subunits are placed according their predicted or known subcellular localizations. Transporters with a blue color background are known to be functional in Y. pestis. On the center/right side, iron storage proteins, the Suf Fe-S cluster assembly system and putative sulfur-mobilizing enzymes (TauD and CysIJ) are displayed. The bottom/right part of the schematic features oxidative stress response proteins. Finally, the top/right part of the schematic displays the flea survival factor Ymt and its fragments, as well as the protein YqhD. These proteins may be implicated in the enzymatic modifications of IM phospholipids. Proteins with a red and green background harbor iron/heme and Fe-S cluster cofactors, respectively.