Table 2. Average log-likelihood for genuine alignments with 500 sequences.
| Method | 16S | COG |
| RAxML 7.0.4 (GTR+CAT or JTT+CAT, SPRs) | −168,104 | −206,724 |
| FastTree 2.0.0 (GTR+CAT or JTT+CAT) | −168,577 | −206,993 |
PhyML 3.0 (GTR+ or JTT+ or JTT+ , no SPRs) , no SPRs) |
−168,603 | −207,156 |
For all topologies, the log likelihood was computed with RAxML 7, re-optimized branch lengths and model parameters, and the GTR+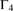 or JTT+
or JTT+ models for 16S or COG, respectively. All differences between FastTree and other methods were statistically significant (
models for 16S or COG, respectively. All differences between FastTree and other methods were statistically significant ( ) except for the comparison with PhyML on 16S rRNAs (
) except for the comparison with PhyML on 16S rRNAs (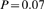 , paired
, paired  test).
test).
