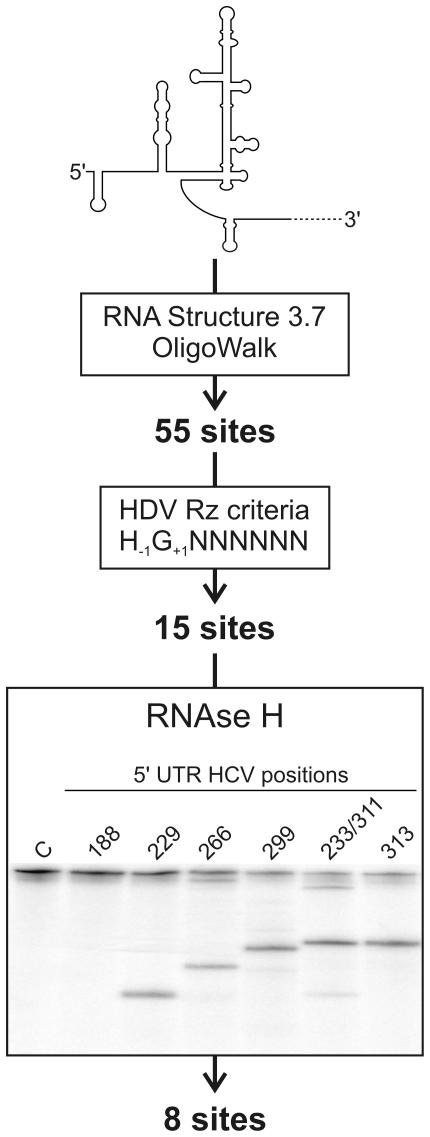
Figure 2. Schematic representation of the 3-step procedure used for the identification of the sites possessing the greatest targeting potential in the HCV 5′-UTR.
Step 1 involved a bioinformatic analysis that included both the prediction of the secondary structure and the identification of the 7 nt streches most likely to be bound by the ribozyme's P1 region using both the RNA structure 3.7 and Oligowalk softwares. Step 2 involved the selection of the sequences that fulfill the HDV ribozyme requirements. Step 3 involved the RNase H hydrolysis assay. The autoradiogram shown corresponds to a typical 5% polyacrylamide gel performed for the analysis of 6 potential sites. The positions of proposed cleavage sites are identified at the top of the gel. The negative control performed in the absence of any oligonucleotide is indicated by the letter C.