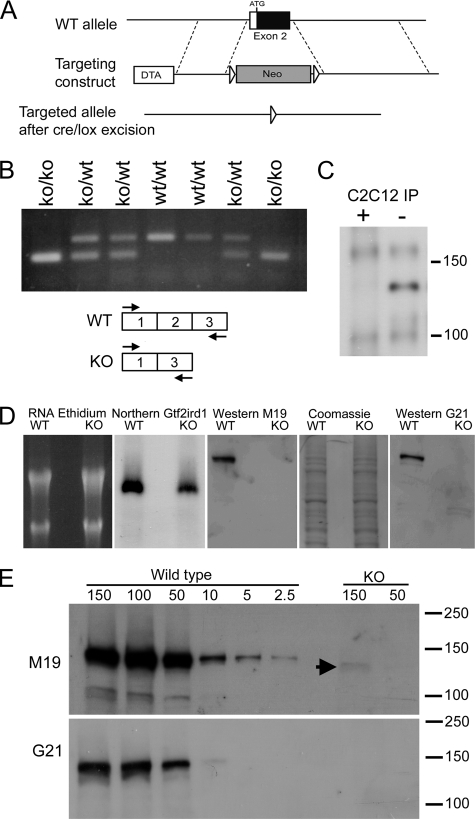
FIGURE 1.
Generation of the Gtf2ird1tm1Hrd mutant allele and the consequences for transcription and translation. A, homologous recombination of the targeting construct with the wild-type (WT) allele using positive neomycin (Neo) selection and negative diphtheria toxin (DTA) selection. The Neo cassette was subsequently removed using the flanking LoxP sites (arrowheads). B, RTPCR analysis of Gtf2ird1 transcripts in brown adipose tissue from a litter of mice segregating the mutant allele using primers in exon 1 and exon 3 (shown in scheme). A novel exon 1–3 splice transcript is produced with equal efficiency to the wild-type transcript. C, Western blot of proteins immunoprecipitated (IP) from C2C12 cell extracts using the M19 antibody for IP and as probe. The immunoprecipitation was conducted in the presence (+) or absence (−) of a blocking peptide antigen to demonstrate specificity. D, analysis of transcription and translation from a wild-type Gtf2ird1 cDNA and the equivalent mutant cDNA lacking exon 2. Extracts from transfected COS-7 cells show that both transcripts are abundantly produced (Northern Gtf2ird1) but the GTF2IRD1 peptide is below detectable levels in cells expressing the mutant transcript (KO) using two antibodies, M19 and G21. Equal loading is shown by ethidium bromide staining of RNA and Coomassie Blue staining of protein in the M19 Western blot. E, Western blot analysis using sequential dilutions of protein extract from cells transfected with the wild-type construct compared with high concentration loadings of mutant extract (KO), probed with M19 and G21 antibodies. A faint band is visible at the highest concentration in the M19 analysis (arrowhead). Numbers represent μg of total protein.