FIGURE 5.
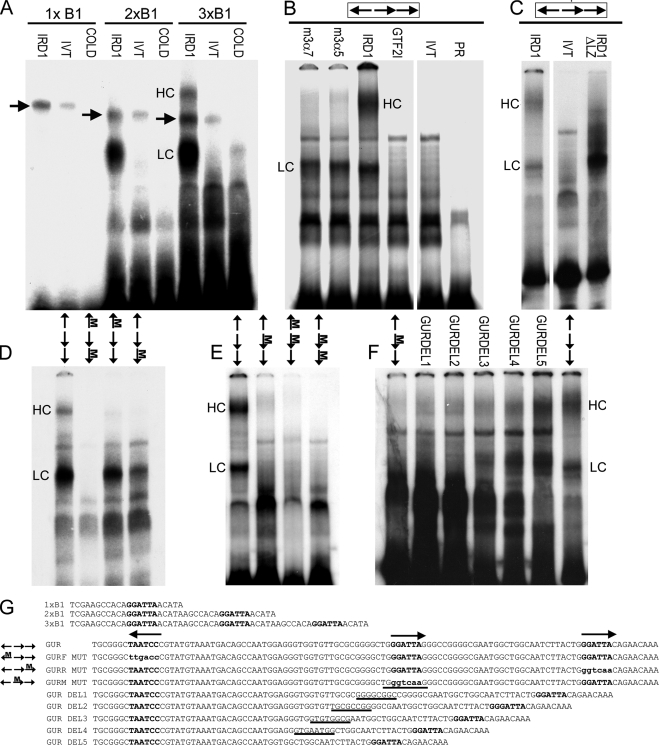
EMSA of GTF2IRD1 proteins showing affinity for the GUR and the tandem nature of the binding. A, binding of human GTF2IRD1 to synthetic multimers of the TNNI1-derived probe, B1, originally used as a yeast one-hybrid bait (10). GTF2IRD1 (IRD1) does not bind detectably to single copies, but binds well to B1 dimers (2xB1) and trimers (3xB1). A lower shift complex (LC) is present in both, but a higher shift complex (HC) appears with the trimer. Probes combined with unprogrammed in vitro translation mix (IVT) or with GTF2IRD1 and excess cold competitor oligonucleotide (COLD) showed no shift. An unknown endogenous shift complex (arrows) present in the in vitro translation mix shows only a minor affinity increase as a result of multimerization. B, GUR fragment is bound efficiently by the two most abundant mouse isoforms of GTF2IRD1, 3α7, and 3α5 and by human GTF2IRD1, but not by the mouse β isoform of TFII-I (GTF2I). No shift occurs in the IVT control or probe-only lane (PR). C, deletion of the leucine zipper domain of GTF2IRD1 (IRD1ΔLZ) intensifies affinity of the LC, but ablates the HC, demonstrating that the HC contains a GTF2IRD1 dimer. D and E, probes containing mutations in single GGATTA sites or in combinations were tested with human GTF2IRD1 protein to determine changes in affinity. Position of the mutation (M) is indicated on the scheme of the GUR (arrows). F, mutation of the middle site alone leads to absence of binding, but the shifts are restored by reducing the distance between the flanking sites, as illustrated using a series of probes with successive deletions (GURDEL1–5). G, sequences of the double-stranded probes used in the EMSA: binding sites are indicated in bold type, and mutations are indicated in lowercase bold type. In the GUR DEL series, the residues deleted in each successive probe are indicated by underlines. Gaps between images indicate where irrelevant lanes have been removed.