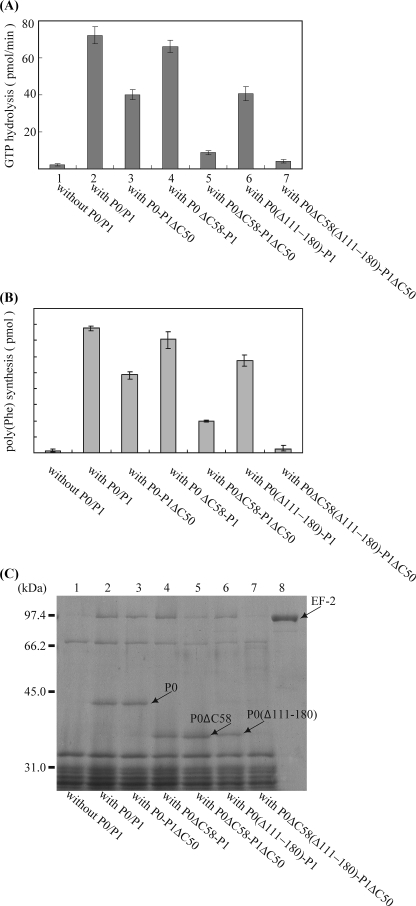
FIGURE 7.
Functional contributions of the C-terminal regions of P1/P0 and of the insertion domain of P0. A, effects of the truncation mutations of the stalk complex on eEF2-dependent GTPase. E. coli 50 S core (2.5 pmol) was preincubated without stalk complex (column 1) or with 20 pmol of the following complexes: column 2, P0–P1 (P0: wild type; P1: wild-type); column 3, P0–P1ΔC50; column 4, P0ΔC58-P1; column 5, P0ΔC58-P1ΔC50; column 6, P0(Δ111–180)-P1; and column 7, P0(Δ111–180)ΔC58-P1ΔC50. The resultant particles were then assayed for eukaryotic eEF2-dependent GTPase activity in the presence of PhL11 (7.5 pmol) and E. coli 30 S subunits (7.5 pmol). B, the same samples (10 pmol of core) as in A were assayed for eukaryotic eEF1α- and eEF2-dependent poly(U)-directed polyphenylalanine synthesis in the hybrid ribosome system (42). C, effects of the truncation mutations of the stalk complex on eEF2 binding. E. coli 50 S core (30 pmol) was preincubated without any stalk complex (lane 1) or with 120 pmol of the following complexes: lane 2, P0–P1; lane 3, P0–P1ΔC50; lane 4, P0ΔC58-P1; lane 5, P0ΔC58-P1ΔC50; lane 6, P0(Δ111–180)-P1; and lane 7, P0(Δ111–180)ΔC58-P1ΔC50. The resultant particles were then incubated with eukaryotic eEF2 (lane 8, eEF2 alone) and GMPPNP, together with PhL11 (90 pmol) and E. coli 30 S subunits (90 pmol), and the eEF2·ribosome complexes were recovered by ultracentrifugation. A given amount of each complex was analyzed by SDS-PAGE. The gels were stained with Coomassie Brilliant Blue.