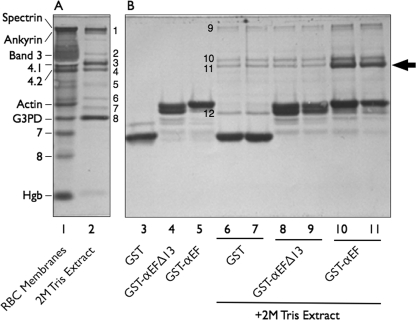
FIGURE 2.
Affinity purification of proteins interacting with the EF-domain of α-spectrin. A, Amido Black-stained 9% Laemmli-Tricine SDS gel of red cell membrane proteins (20 μg) and the protein composition of a 2 m Tris extract (5 μg) of spectrin- and actin-depleted membranes are shown. Protein bands (B) present in the extract were determined by mass spectrometry: B1, ankyrin; B2, unidentified; B3, protein 4.1; B4, protein 4.2; B5, protein p55; B6, dematin; B7, aldolase A; B8, glyceraldehyde-3-phosphate dehydrogenase (G3PD). RBC, red blood cells. Hgb, hemoglobin globin chains. B, shown is analysis of recombinant GST alone and GST-EF-domain fusion proteins retrieved by GSH beads before (lanes 3 to 5) and following (lanes 6 to 11) incubation with a 2 m Tris membrane protein extract that had been dialyzed into binding buffer. The gel is a separate section of the gel shown in A. Eight μg of each sample were applied to the gel with the exception of lanes 6, 8, and 10, where 16 μg were used. Protein bands B9-B12 were determined by mass spectrometry: B9, ankyrin; B10, protein 4.1; B11, protein 4.2; B12, glyceraldehyde-3-phosphate dehydrogenase. Note that the intact EF-domain (GST-αEF) retrieves protein 4.2 (arrow) from the extract, but the same domain is inactive if the last 13 amino acids are deleted (GST-αEFΔ13).