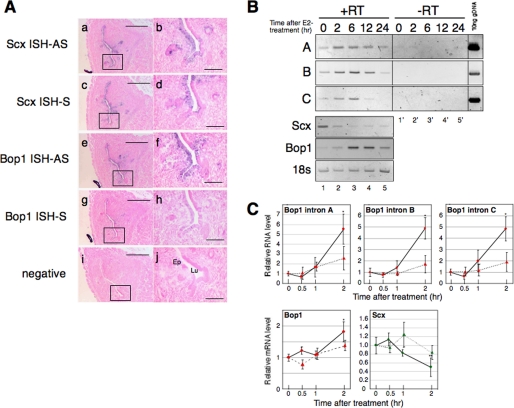
FIGURE 3.
Antisense RNA for the Scx gene transcript exists in the OVX mouse uterus. A, ISH results from serial sections of the OVX mouse uterus. ISH was performed using an antisense probe (AS) for Scx cDNA (panels a and b), a sense probe (S) for Scx cDNA (panels c and d), an antisense probe for Bop1 cDNA (panels e and f), and a sense probe for Bop1 cDNA (panels g and h). The position of the probes is indicated in Fig. 1. A nonspecific plant sequence was used as the negative control (panels i and j) as described under “Experimental Procedures.” The purple signal shows specific hybridization of the indicated probes. The boxes in panels a, c, e, g, and i are increased in magnification correspond to panels b, d, f, h, and j, respectively. Ep indicates endometrial epithelial cells, and Lu indicates lumen. Scale bars = 200 μm in panels a, c, e, g, and i and 50 μm in b, d, f, h, and j. B, total uterine RNA was prepared from the E2-treated mice at the indicated times. A representative RT-PCR result of various primer sets (A, B, C, Scx, Bop1, and 18 S) using the identical samples is shown. A, B, and C indicate the Bop1 intronic elements in Fig. 1. The samples of lanes 1–5 were reverse-transcribed from RNA extracted from E2-treated uteri at the indicated times (+RT). The samples of lanes 1′–5′ were not reverse-transcribed using the same RNA of 1–5 (−RT). Ten ng of genomic DNA were used as the positive control for the intronic RNA. C, E2 was administered to mice (six mice per group) with (dashed line) or without (solid line) ICI, and uterine total RNA was prepared at the times indicated. Real-time PCR was performed, and the results were normalized to the level of 18 S in each sample. The values are the means ± S.D. *, p < 0.01 when compared with time 0, **, p < 0.02 when compared with time 0.