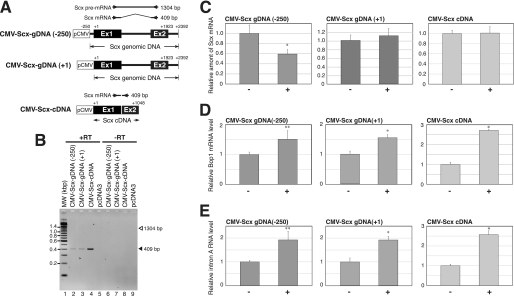
FIGURE 5.
Scx mRNA expressed from the exogenous Scx genomic DNA is reduced by increasing the endogenous Bop1 gene expression. A, schematic representation of CMV-Scx-gDNA(+1), CMV-Scx-gDNA(−250), and CMV-Scx-cDNA is shown. The position of PCR primers used for the amplification of Scx mRNAs is indicated. The expected size of the PCR amplicons is shown to the right of each primer pair. Ex1 and Ex2 indicate exons 1 and 2, respectively. B, a representative RT-PCR result of Scx transcript is shown. The samples were prepared from BALB/3T3 cell lines containing CMV-Scx-gDNA(+1), CMV-Scx-gDNA(−250), CMV-Scx-cDNA, or pcDNA3 (as a negative control). The samples in lanes 2–5 were reverse-transcribed (+RT) from the RNA, and the samples of lanes 6–9 were not reverse-transcribed (−RT) using the same RNA of 2–5. MW, molecular weight markers. C, total RNA was prepared from treated (high serum medium; +) or untreated (low serum medium; −) CMV-Scx-gDNA(+1), CMV-Scx-gDNA(−250), and CMV-Scx-cDNA BALB/3T3 cells as described under “Experimental Procedures.” A representative real-time PCR result of Scx mRNA is shown. D, a representative real-time PCR result of Bop1 mRNA is shown. E, a representative real-time PCR result of Bop1 intronic RNA (primer set A) is shown. The RNA level was normalized to the level of 18 S in each sample. The values are the means ± S.D. for three independent replicates. *, p < 0.01 when compared with (−), **, p < 0.05 when compared with (−).