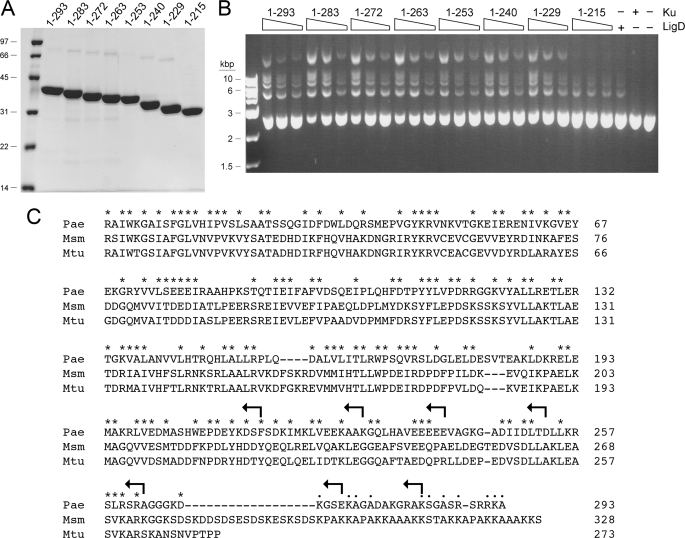
FIGURE 5.
Ku stimulates LigD ligase activity. A, nickel-agarose preparations of full-length Ku-(1–293) and the indicated KuΔ mutants were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is show. The positions and sizes (kDa) of marker polypeptides are indicated on the left. B, reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 5 mm MnCl2, 0.25 mm ATP, 0.2 μg of KpnI-digested pUC19 DNA (∼115 fmol of plasmid; 230 fmol of double strand breaks), 0.1 μg (1 pmol) of PaeLigD, and 30, 60, or 120 ng (from right to left in each titration series) of full-length Ku or KuΔ mutants as specified were incubated for 30 min at 37 °C. Control reaction mixtures contained 0.1 μg of LigD alone, 120 ng of Ku alone, or neither protein. The reactions were quenched by adjusting the mixtures to 0.2% SDS, 6 mm EDTA. The products were resolved by electrophoresis through a horizontal 1% agarose gel containing TBE (90 mm Tris borate, 2.5 mm EDTA) and 0.05% ethidium bromide. A photograph of the gel under UV illumination is shown. The positions and sizes (kbp) of linear duplex DNA markers are indicated on the left. C, amino acid sequences of the Ku polypeptides of P. aeruginosa (Pae), M. smegmatis (Msm), and M. tuberculosis (Mtu) are aligned. Positions of side chain identity or similarity in all three proteins are indicated by an asterisk. Gaps in the alignment are indicated by a dash. The C termini of the KuΔ mutants are indicated by arrows.