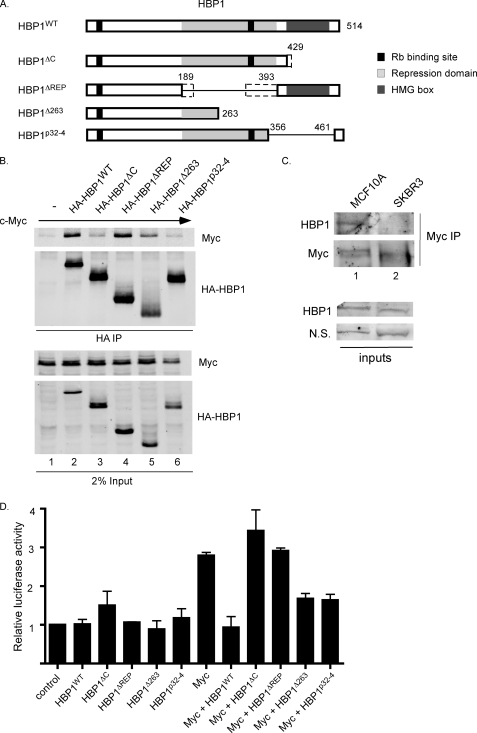
FIGURE 5.
c-Myc shows reduced binding to and/or altered activity in the presence of some HBP1 mutants. A, shown are schematics of HBP1 and HBP1 deletion mutants used in co-immunoprecipitations. HBP1Δ263 and HBP1p32-4 have been previously described as being isolated from human breast tumors (35). Other mutants isolated in breast cancer that are similar to deletion mutants generated for this study are indicated by dotted lines; specifically, a mutant C-terminal truncated at amino acid 431 and a mutant deleted for amino acids 218–314. B, c-Myc binds weakly to the HBP1 C-terminal deletion mutant and has reduced binding to some HBP1 breast cancer-derived mutants. 293 cells were transiently transfected with CMV-β-gal, V5-c-Myc, and either empty vector or one of the indicated HA-HBP1 expression vectors. Cleared lysates were normalized to β-galactosidase and subjected to immunoprecipitation with anti-HA. Inputs and immunoprecipitated complexes were analyzed by Western blotting with the indicated antibodies. C, the interaction between HBP1 and c-Myc is not detected in SKBR3 breast cancer cells. The indicated cells were lysed in co-IP buffer, and c-Myc was immunoprecipitated with C33-conjugated protein A-Sepharose. Immunoprecipitation was visualized by Western blotting with c-Myc (N262) and HBP1 antibodies. Inputs are shown to demonstrate similar HBP1 expression levels in MCF10A and SKBR3 cells. D, some HBP1 mutants fail to repress c-Myc-induced transcription from a synthetic E-box promoter. 293 cells were transiently transfected with either control pGL2 or 4xEbox-luciferase and CMV-β-gal together with V5-c-Myc and/or various HA-HBP1 expression vectors as indicated. 18–20 h post-transfection, cells were serum-starved in DMEM supplemented with 0.2% serum for 48 h. Cleared lysates were analyzed for β-galactosidase and luciferase activity. Luciferase activity was normalized to β-galactosidase activity and is shown relative to control pGL2 activity. The averages of three independent experiments ± S.E., as calculated in GraphPad Prism, are shown.