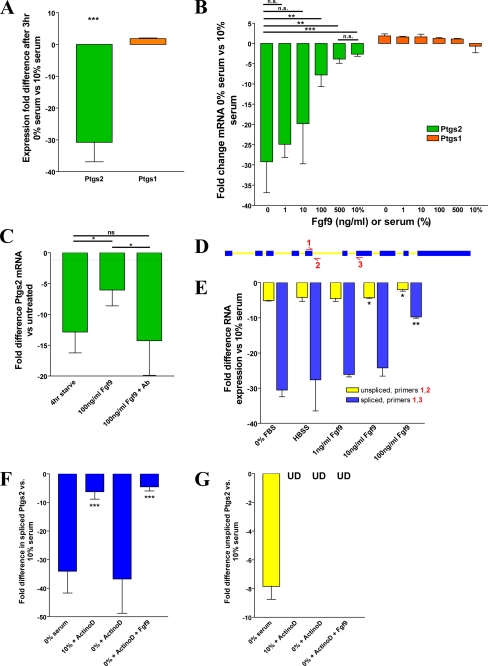
FIGURE 6.
Fgf9 is sufficient to stabilize Ptgs2 mRNA. qRT-PCR analysis (A–C and E–G) of cMSCs is shown. A, graph of the -fold difference for Ptgs2 and Ptgs1 expression of serum-starved cMSCs versus cMSCs grown in 10% serum. B, graph of the -fold difference for Ptgs2 and Ptgs1 expression in cMSCs after 1 h of Fgf9 treatment (range = 1–500 ng/ml) following 3 h of serum starvation. C, graph of the -fold difference for Ptgs2 expression after treatment with Fgf9 or Fgf9 plus anti-Fgf9 neutralizing antibody following 3 h of serum starvation. D, schematic illustrating the location of primers 1, 2, and 3 on the Ptgs2 gene. E, graph of the -fold difference for the spliced and unspliced forms of Ptgs2 isolated from 3-h serum-starved cMSCs additionally treated for 1 h with Fgf9. F, graph showing the -fold difference for Ptgs2 spliced RNA after treatment with or without serum starvation, 4 μg/ml actinomycin D, and 250 ng/ml Fgf9. The asterisks show comparison of data with -fold difference in cells treated with 0% serum and actinomycin D. G, graph showing the -fold difference for Ptgs2 unspliced RNA with or without serum starvation, 4 μg/ml actinomycin D, and 250 ng/ml Fgf9 (UD, undetectable). All data are representative of three independent experiments. Error bars, S.D. Statistical analysis by Student's t test is shown: *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.