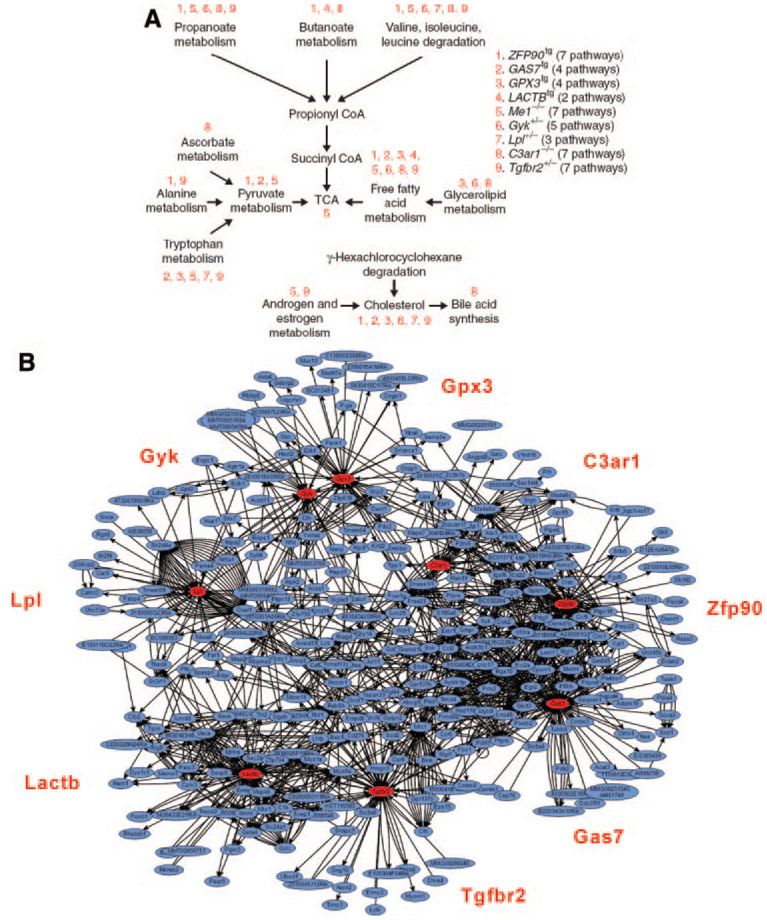
Figure 2.
Networks contributing to adiposity in mice. A, A set of curated pathways was shown to be enriched in genes differentially expressed between livers of fat and lean mice in a segregating mouse population.9 Note that these pathways tend to center on the tricarboxylic acid cycle, which is central in energy production. Through the use of systems genetics, a set of genes (labeled 1 through 9) were predicted to be causally related to adiposity in mice.10 Their effects on adiposity were validated with transgenic approaches, and expression array analyses of the transgenic mice showed that the differentially regulated genes were enriched in many of the same tricarboxylic acid–centered pathways (indicated by numbering above pathways).11 B, Systems genetics approaches were used to construct a directed “bayesian” coexpression network model based on global expression array analysis of segregating populations of mice. One subnetwork, or module, was significantly associated with adiposity. Notably, the genes causally involved in adiposity (above) correspond to hubs in the network. Data derived from refs. 9, 11.