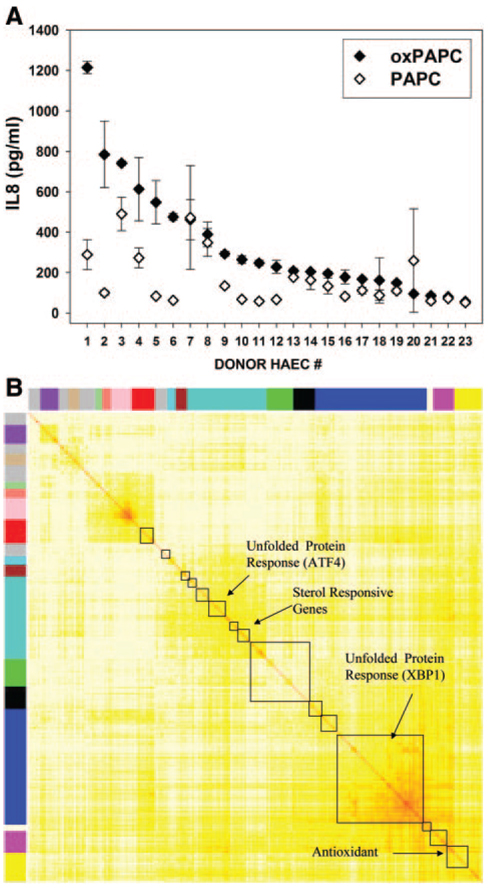
Figure 6.
Coexpression analysis of the inflammatory responses of human endothelial cells based on common genetic variations in the population. Small pieces of human aortic arteries (obtained during the course of heart transplant surgery) were used to obtain pure, early-passage cultures of human endothelial cells. The endothelial cells from different donors were observed to exhibit significant differences in response to oxidized lipids. A, Production of interleukin-8 (IL8) by the cultures after treatment with biologically active oxidized phospholipids (solid symbols) vs control levels (open symbols). A total of 12 endothelial cell cultures isolated from individuals exhibiting various responses to oxidized lipids were subjected to microarray analysis, and >1000 genes were shown to be regulated by oxidized lipid treatment. Transcript levels of these genes were then used to model scale-free networks based on coexpression. A correlation matrix of all the genes was constructed, and their connectivities were used to generate a topographical overlap matrix (TOM) plot. B, Results shown as color-coded clusters (modules) of highly correlated genes in a symmetric plot. Correlations between genes are indicated by the color intensity (white corresponding to little or no correlation; red, strong correlation). The red diagonal represents correlations of each gene with itself, and offset from this are correlations with other genes. Some of the modules were highly enriched for curated pathways as shown. Adapted from Gargalovic et al32 according to the policy of Proceedings of the National Academy of Sciences. Copyright © 2006 by the National Academy of Sciences. HAECs indicates human aortic endothelial cells.