Abstract
The need for selective matrix metalloproteinase (MMP) inhibition is of interest because of the range of pathologies mediated by different MMP isoforms. The development of more selective MMP inhibitors (MMPi) may help to overcome some of the undesired side effects that have hindered the clinical success of these compounds. In an effort to devise new approaches to selective inhibitors, herein we describe several novel MMPi and show that their selectivity is dependent on the nature of the zinc-binding group (ZBG). This is in contrast to most current MMPi, which obtain isoform selectivity solely from the peptidomimetic backbone portion of the compound. In the present study, six different hydroxypyrone and hydroxypyridinone ZBGs were appended to a common biphenyl backbone and the inhibition efficiency of each inhibitor was determined in vitro (IC50 values) against MMP-1, -2, -3, -7, -8, -9, -12, and -13. The results show that the selectivity profile of each inhibitor is different as a result of the various ZBGs. Computational modeling studies were used to explain some trends in the observed selectivity profiles. To assess the importance of the ZBG in a biological model, two of the semiselective, potent MMPi (and one control) were evaluated using an isolated perfused rat heart system. Hearts were subjected to ischemia reperfusion injury, and recovery of contractile function was examined. In this model, only one of the two MMPi showed significant and sustained heart recovery, demonstrating that the choice of ZBG can have a significant effect in a relevant pathophysiological endpoint.
Keywords: bioinorganic chemistry, extracellular matrix, ischemia-reperfusion, matrix metalloproteinase inhibitor, zinc
Introduction
Matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases involved in the breakdown of the extracellular matrix (ECM) and basement membrane components such as aggrecan, collagen, elastin, fibronectin, gelatin, and laminin.[1–3] In normal physiology, MMPs are responsible for ECM homeostasis, tissue remodeling, wound healing, angiogenesis, and apoptosis amongst other important processes.[1, 4–6] However, MMPs can also participate in the pathophysiology of diseases such as arthritis, multiple sclerosis, periodontal disease, cancer metastasis, atherosclerosis, and cardiac injury and remodeling.[7–12] MMPs have therefore been an important therapeutic target for over two decades, with growing interest to-wards the design of inhibitors that are highly specific against different MMP isoforms and possess a high therapeutic index over older generation compounds.[13–19]
Although there is considerable substrate overlap amongst the different MMPs, these enzymes have often been classified based on their substrate specificity into gelatinases (MMP-2, -9), collagenases (MMP-1, -8, -13), stromelysins (MMP-3, -10), membrane type (MT-MMPs, MMP-14, -15, -16, -17), matrilysin (MMP-7), metalloelastase (MMP-12), and unclassified.[5, 18, 19] These multidomain proteins have certain domains in common, which include the signal domain, the propeptide domain, and the catalytic domain.[12, 14, 17–21] The catalytic domain of all MMPs contain a Zn2+ ion coordinated by a tris(histidine) motif; the Zn2+ ion is critical for both substrate binding and cleavage.[12, 14, 17, 21] Most MMP inhibitors (MMPi) consist of two parts: a zinc-binding group (ZBG) to bind the catalytic metal ion[13, 14, 21] and a peptidomimetic backbone to interact noncovalently with specific subsites neighboring the active site of the protein.[16, 17, 22] The catalytic Zn2+ in the active site is surrounded by subsite pockets designated as S1, S2, S3, S1′, S2′, and S3′.[23, 24] Of the different subsite pockets, targeting of the S1′ pocket has provided the basis of selectivity for many MMPi.[15, 23] As will be discussed below, in the current study, a hydrophobic biphenyl backbone was used to select against shallow S1′ pocket MMPs, such as MMP-1 and MMP-7.[14, 23, 25]
The most common ZBG used in MMPi is the hydroxamic acid group, which has produced numerous nanomolar inhibitors, but has not been successful in clinical trials.[20, 23, 26] The failure of hydroxamic acid MMPi at a clinical level may stem, in part, from the lack of selectivity of hydroxamic acids towards the Zn2+ ion, poor pharmacokinetics, and poor oral bioavailability. Consequently, some efforts have been made to identify alternatives to the hydroxamic acid ZBG. Most of these studies have only compared hydroxamic acids to carboxylic acids (the synthetic precursor to most hydroxamic acids) and generally observed a loss in potency,[26, 27] likely due to the change in binding mode and diminished donor ability of the carboxylate ligand.[18] However, Castelhano and co-workers reported the selectivity and potency of MMPi with several different ZBGs (for example, hydroxamates, ’reverse’ hydroxamates, carboxylates, thiols, phosphinates) on a common indolactam/isobutyl backbone moiety.[28] The results of this study showed that the use of ZBGs other than a hydroxamate resulted in a 10- to 250-fold loss in potency. Although several MMPs were tested, no extensive study of isoform selectivity was performed, but the data available generally showed no changes in selectivity based on the nature of the ZBG. Another study by Fesik and co-workers used SAR-by-NMR (structure–activity relationship by nuclear magnetic resonance) to screen several molecular fragments to identify new ZBGs; ultimately a naphthyl-substituted hydroxamic acid was found to be the most potent ZBG, but no other systematic studies were performed.[22] Other efforts have been made toward the identification of alternative ZBGs using a novel bioinorganic approach.[18, 29–33] These studies have identified new ZBGs, that are more potent than hydroxamic acids, some of which have been developed into potent, nonhydroxamate inhibitors of MMPs.[32, 34, 35] In the present study, a series of hydroxypyrone and hydroxypyridinone inhibitors have been synthesized and evaluated to demonstrate that the isoform selectivity of an MMPi can be influenced by the choice of ZBG. These observations are particularly notable, because the ZBGs employed in this study all use the same type and number of donor atoms (two oxygen atoms) to bind the Zn2+ ion, and only possess subtle differences in electrostatics, hydrophobicity, and acidity. This is an unprecedented finding, as previous studies have focused only on the backbone substituents to obtain selective MMP inhibition.[15, 23, 36]
To examine isoform selectivity, the compounds were evaluated in vitro against eight MMPs using a fluorescent peptide substrate-based assay. The results indicate that both the ZBG and the backbone contribute to selective MMP inhibition. By linking different ZBGs to a common biphenyl backbone, we demonstrate that small changes in the ZBG modulate the selectivity against deep S1′ pocket MMPs. Furthermore, to demonstrate the importance of the ZBGs in a relevant pathophysiological model, we tested two MMPi and one control compound (each with a different ZBG) using an isolated perfused rat heart (Langendorff) system. Cardiac myocytes and other myocardial cells express multiple MMPs which are involved in organ ECM homeostasis.[7, 8, 37] Ischemia-reperfusion (IR) injury in ex vivo heart preparations is known to activate MMPs that mediate myocardial damage, and as a consequence, loss of contractile function.[38] Measurements of the recovery of contractile performance with different MMPi after a period of global IR demonstrate that the ZBG significantly impacts the magnitude of recovery of function. Taken together, the in vitro, computational, and ex vivo results described herein show that MMPi presented here display different isoform selectivity and varying therapeutic potentials depending on the ZBG employed.
Results
Synthesis of MMPi
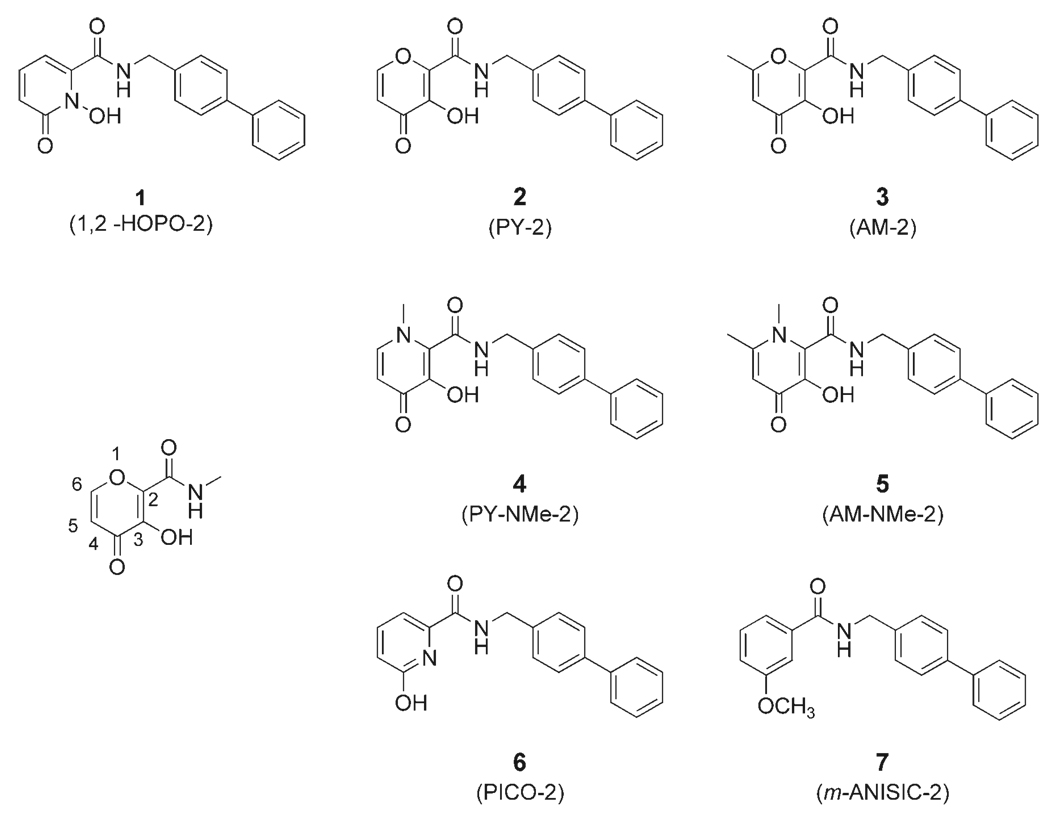
Previous work from our laboratory showed that several hydroxypyrone and hydroxypyridinone ZBGs had improved potency against MMPs when compared to a standard hydroxamate compound, acetohydroxamic acid (AHA).[30, 31] A novel, full-length inhibitor AM-2 (compound 3, Figure 1), based on a hydroxypyrone ZBG, was shown to be a potent inhibitor of MMP-2 and MMP-3 but not MMP-1.[32] These results showed that potent, nonhydroxamate MMPi could be devised, and AM-2 was selected as the basis for the design and synthesis of the MMPi examined here (Figure 1). Each inhibitor contains a chelating, heterocyclic ZBG attached to the same biphenyl backbone as in AM-2 (see Supporting Information for synthetic details). Generally, the inhibitors were prepared by benzyl protection of the hydroxy group of each ZBG, followed by conversion of each benzyl-protected ZBG from a carboxylic acid to an activated ester, which were then coupled with phenylbenzylamine, and finally deprotected to obtain the compounds shown in Figure 1. The MMPi in Figure 1 are relatively facile to prepare and readily allow for preparation in multigram scales. As negative controls, compounds 6 and 7 were synthesized, both of which contain the same biphenyl backbone as the other inhibitors, but use an isosteric (6) or nearly isosteric (7) ring that is not an effective Zn2+ chelator. Interestingly, the ZBG (6-hydroxypicolinic acid)[39] of compound 6 is the synthetic precursor to the chelating ZBG used in compound 1.
Figure 1.
Structures of the inhibitors examined in this study (1–7). An abbreviated name for each inhibitor is provided in parentheses. Compounds 6 and 7 are negative controls, as they do not contain a high affinity ZBG. The numbering scheme used for the pyrone ring system is shown on the left.
In vitro efficacy of inhibitors
Inhibitors 1–7 were tested against several human MMP catalytic domains using commercially available kits that employ a fluorogenic substrate, with observed fluorescence directly proportional to MMP activity.[40] IC50 values were determined (Table 1) as previously described,[31, 33, 40] and enzyme kinetic experiments were performed to determine the Km value, Ki value, and the mode of inhibition. As expected, MMPi 1–5 are poor inhibitors of shallow S1′ pocket MMPs (that is, MMP-1 and MMP-7) with IC50 values greater than 50 µm. The MMPi also show poor inhibition against MMP-9 with IC50 values greater than 25 µm. MMP-13 is inhibited with greater potency (IC50 values ~4–20 µm) by MMPi 1–5 with no notable differences in potency based upon the different ZBGs. MMP-2 is inhibited by compound 1 with a nanomolar IC50, but compounds 2–5 display inhibition profiles similar to that of MMP-13 (low micro-molar). MMP-8 and MMP-12 are strongly inhibited by compounds 1–3 with IC50 values ranging from 0.018–0.248 µm, but are inhibited with lesser potency by 4 and 5 (1.2–5 µm). Similarly, inhibitors 1–3 show strong potency against MMP-3 with IC50 values of 0.56, 0.077, and 0.24 µm respectively. In contrast, the corresponding hydroxypyridinone analogues 4 and 5 were more than two orders of magnitude less potent against MMP-3. As a control, compounds 6 and 7 were also tested against MMP-8 and MMP-12 and showed no or negligible inhibition at concentrations up to 100 µm. The weak activity of compounds 6 and 7 clearly demonstrates that a tight binding ZBG is required for potent inhibition and that the biphenyl backbone alone is not sufficient for efficacy. Collectively, the results show that the nature of the ZBG has a pronounced effect on the scope and potency of the resulting MMPi.
Table 1.
IC50 values for MMPi 1–7 (rows) against eight different MMPs (columns) using a fluorescence-based assay.[a]
| MMP | −1 | −2 | −3 | −7 | −8 | −9 | −12 | −13 |
|---|---|---|---|---|---|---|---|---|
| 1 | >50 | 0.92 (0.04) | 0.56 (0.042) | >50 | 0.086 (0.009) | 27.1 (5.3) | 0.018 (0.001) | 4.1 (0.6) |
| 2 | >50 | 4.4 (1.4) | 0.077 (0.005) | >50 | 0.248 (0.005) | 32.3 (2.5) | 0.085 (0.004) | 6.6 (0.1) |
| 3 | >50 | 9.3 (0.5) | 0.24 (0.01) | >50 | 0.064 (0.008) | >50 | 0.022 (0.002) | 20.6 (3.0) |
| 4 | >50 | 16.5 (2.8) | 41.7 (4.8) | >50 | 3.8 (0.1) | >50 | 1.2 (0.4) | 16.5 (3.0) |
| 5 | >50 | 7.6 (0.2) | >50 | >50 | 5.0 (0.4) | >50 | 6.7 (1.5) | 6.7 (0.2) |
| 6 | ND | ND | ND | ND | >100 | ND | >100 | ND |
| 7 | ND | ND | ND | ND | >100 | ND | >100 | ND |
Compounds 6 and 7 showed no inhibition at concentrations up to 100 µm. Values in parentheses are standard deviations of at least four independent measurements. ND=Not determined. IC50 values are given in µm.
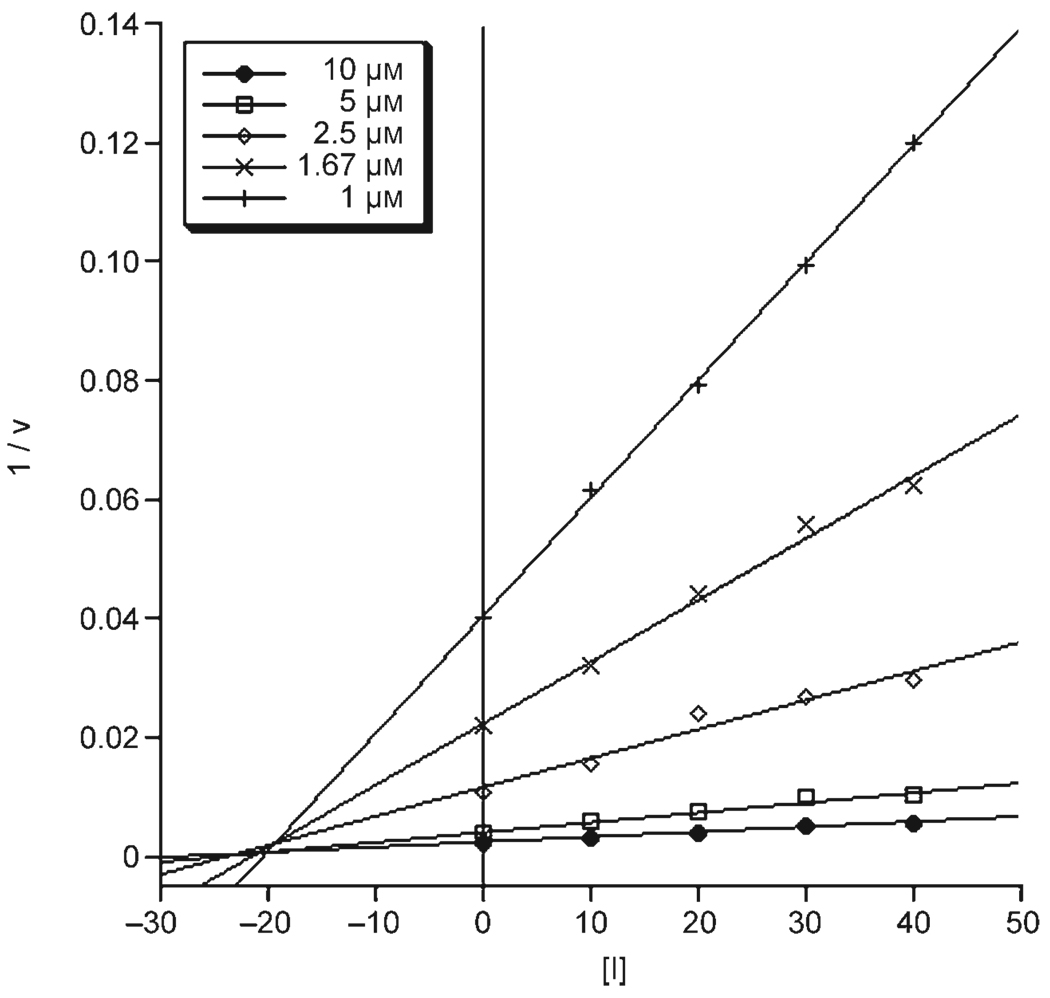
As a representative example, compound 1 was selected for further kinetic studies against MMP-12 (the isoform against which 1 is most potent). Kinetic studies were performed using the same fluorescence-based assay kit used for IC50 measurements with identical ratios of enzyme, substrate, and inhibitor. Estimations of Km were obtained from plots of reaction velocity versus substrate concentration assuming a Michaelis–Menten mechanism, using substrate concentrations ranging from 0 to 80 µm. The Vmax and Km for MMP-12 were determined to be 829 mmol min−1mg−1 and 4 µm, respectively. The value of Ki for 1 against MMP-12 was calculated from equation 1 (vide infra) to be 9 nm.[41] A Dixon plot of the reciprocal of initial velocity (calculated from equation 2) versus inhibitor concentration, with concentrations of inhibitor ranging from 0 to 40 nm, was plotted to determine the mode of inhibition and confirm the calculated Ki value. The Dixon plot shown in Figure 2 is indicative of a mixed competitive mode of inhibition with an experimental Ki value of approximately 18 nm,[41, 42] which is consistent with the observed IC50 value.
Figure 2.
Dixon plot of MMP-12 with different substrate concentrations (10 µm=filled circles, 5 µm=open squares, 2.5 µm=open diamonds, 1.67 µm=crosses, 1 µm=plus signs) against varying concentrations of MMPi 1 (1/v in mmol−1min mg, [I] in nm).
Ex vivo efficacy of inhibitors
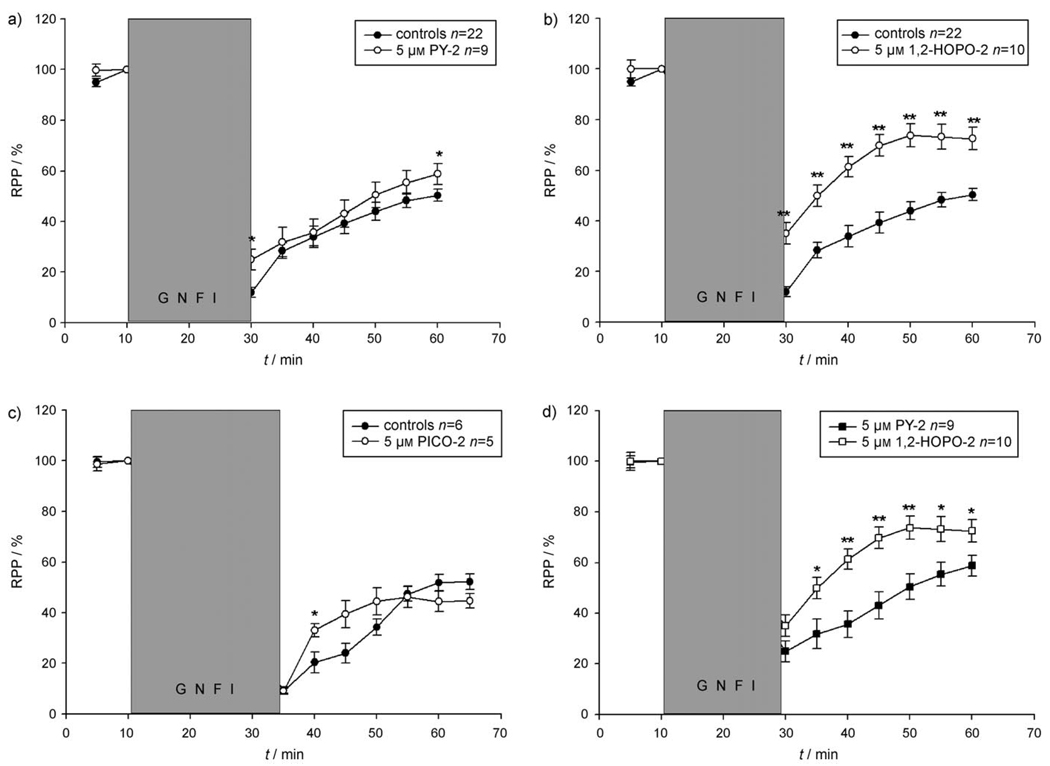
An ex vivo rat heart model was used to provide a biologically relevant system for examining the effects of MMPi containing different ZBGs. The results from these studies are shown in Figure 3. After implementation of a 20–25 min period of global no-flow ischemia, reperfusion of untreated hearts led to a progressive recovery of contractile function of ~50% of the work index rate-pressure product (RPP) after 30 min reperfusion. This ~50% recovery provided a benchmark to which hearts treated with an MMPi could be compared.
Figure 3.
Recovery of rate pressure product (RPP) in isolated rat hearts subjected to global no-flow ischemia (GNFI) and 30 min reperfusion in the absence (controls) and presence of: 5 µm PY-2 (compound 2, upper left), 5 µm 1, 2-HOPO-2 (compound 1, upper right), and 5 µm PICO-2 (compound 6, lower left). A direct comparison of the recovery results from PY-2 and 1, 2-HOPO-2 are shown in the panel at the lower right. Note, at equal concentrations, 1, 2-HOPO-2 confers a higher recovery of contractile function than PY-2 during reperfusion. All MMPi were present in the perfusion buffer throughout the experiment. *P<0.05, **P<0.01.
Addition of 5 µm of MMPi 2 (PY-2) to the model system led to essentially no improvement in recovery of contractile function when compared with the untreated controls. More significantly, the use of 5 µm of MMPi 1 (1,2-HOPO-2) led to a notable and persistent improvement in the recovery of contractile function (RPP) at all time points after reperfusion (Figure 3). Figure 3 directly compares the values for the recovery of contractile function between hearts treated with compound 1 and 2. MMPi 1 consistently led to a greater improvement in recovery of contractile function when compared to 2. It is important to note that these compounds are entirely isosteric, have nearly identical solubilities, and only differ in the nature of the heterocyclic ZBG (Figure 1). Use of 5 µm of control compound 6 (PICO-2) in the same ex vivo model again yielded essentially no notable differences to the untreated control hearts. The latter result shows that compounds lacking a potent ZBG do not show any recovery effects in this model.
Discussion
Selectivity beyond the backbone
The MMP active site is mostly solvent exposed and is characterized by six subsite pockets as previously mentioned. Apart from substrate classification, MMPs can also generally be described as being shallow, intermediate, or deep pocket enzymes based on the size of their S1′ pocket.[25] MMP-1 and MMP-7 are considered shallow pocket MMPs; MMP-2, MMP-9, and MMP-13 are classified as intermediate pocket MMPs; MMP-3, MMP-8, and MMP-12 are designated deep pocket MMPs.[25] Figure 4 provides a summary of the selectivity for each MMPi against the aforementioned MMPs. The poor IC50 values of all the inhibitors against MMP-1 and MMP-7 are consistent with the description of these enzymes as shallow S1′ pocket MMPs. All the inhibitors tested use a large biphenyl backbone that is incompatible with the small S1′ subsite in these MMPs.[32] The poor inhibition of the compounds against MMP-9 may also be a result of its relatively shallow S1′ subsite being unable to accommodate the bulky biphenyl moiety. These findings are consistent with the known structural data for the MMPs examined, and show the typical selectivity expected based on incorporation of a large S1′-directed backbone substituent.[23]
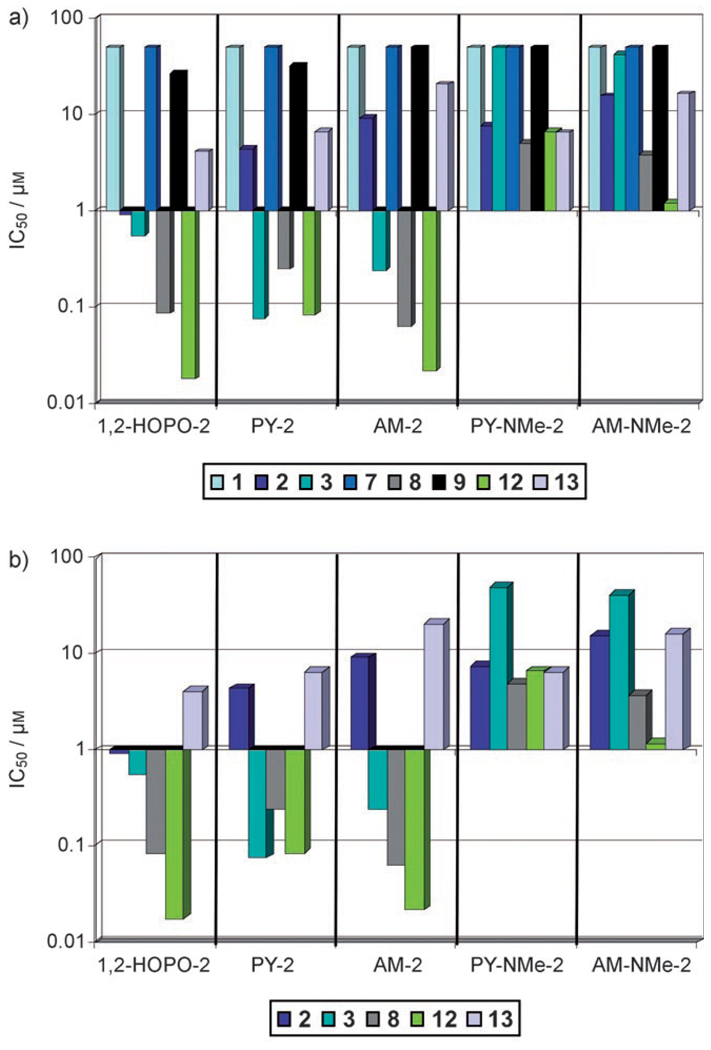
Figure 4.
Graph of IC50 values (on y-axis) from Table 1 plotted on a logarithmic scale for inhibitors 1–5. Each MMP is color coded as shown in the legend (MMP-2 dark blue; MMP-3 turquoise; MMP-8 gray; MMP-12 green; MMP-13 mauve). The top plot shows all of the MMPs tested and the bottom plot shows only the intermediate and deep S1′ pocket MMPs.
Looking beyond the S1′ pocket, examination of the data in Figure 4 reveals that in addition to the selectivity obtained by the backbone, a second level of selectivity (that is, “fine tuning”) was observed based on differences in the ZBGs. When inspecting the inhibition of each compound against MMP-2, we see that 1 shows nanomolar potency, and is almost an order of magnitude better than the other MMPi. The remaining MMPi (2–5) inhibit MMP-2 with IC50 values in the low-micromolar range. With an invariant backbone among these inhibitors, the enhanced potency of 1 against MMP-2 can be attributed to its unique 1-hydroxypyridin-2(1H)-one ZBG. When examining only the subset of MMPs for which compound 1 displays some potency, a >50-fold selectivity for MMP-12 over MMP-2 is observed. Further, compound 1 is at least 2500-fold more potent against MMP-12 over MMP-1 or MMP-7. By comparison, the first generation hydroxamic acid MMPi Batimastat (BB-94) inhibits MMP-1, -2, -3, -8, -9, and -13 with little selectivity, inhibiting all these isoforms within a narrow range of IC50 values (~1–20 nm).[4]
The effect of the ZBG becomes more pronounced in the deep pocket MMPs, MMP-8, and MMP-12. Figure 4 shows that both MMP-8 and MMP-12 are strongly inhibited by MMPi 1–3 with IC50 values in the nanomolar range, but to a much lesser extent by MMPi 4 and 5 with IC50 values in the low-micromolar range. The only structural difference in the ZBGs from 4 and 5 when compared with MMPi 1–3 (Figure 1) is an additional methylated nitrogen atom in the heterocyclic ring system. As expected, MMP-8 and MMP-12 show negligible inhibition by negative controls 6 and 7 (Table 1).
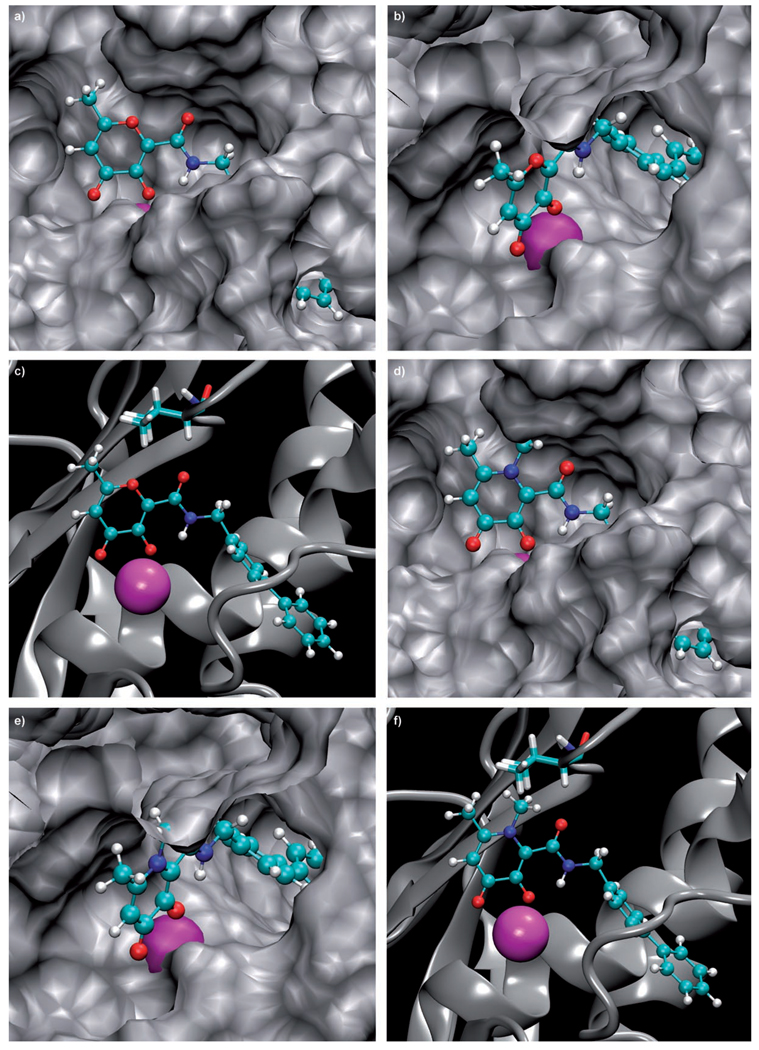
The attenuated potency of the inhibition profiles of 4 and 5 is most prominent against the deep S1′ pocket MMP-3. The N-methyl substituent on MMPi 4 and 5 decreases MMP-3 inhibition by three orders of magnitude compared to their pyrone analogues 2 and 3. Based on computational modeling, the decrease in inhibition caused by the methyl substituent can be attributed to a steric clash within the active site of MMP-3 as shown in Figure 5. The region responsible for the steric conflict lies directly above the catalytic Zn2+ ion. The N-methyl group on MMPi 4 and 5 collides with a methyl group on the isopropyl side chain of Val163 in MMP-3, as confirmed by performing a ’bump’ analysis using Insight II (Accelrys). Interestingly, residue 163 is also a significant residue in the S1 subsite of different MMP isoforms, where it frequently is found to interact with inhibitors targeting the S1 subsite. The variability in residue 163 has been exploited in the design of inhibitors to confer selectivity against different MMPs.[23, 43] In contrast, Figure 5 also shows that methyl substituents in the 6-position of the pyrone ring (for example, compound 3, Figure 1) do not lead to any steric conflicts within the MMP-3 active site. Indeed, this methyl group appears to contribute to the isoform selectivity observed for MMPi 3.
Figure 5.
Three different views comparing the binding of MMPi 3 (left) and 5 (right) in the MMP-3 active site. Zn2+ ions are shown as magenta spheres, the MMPi in ball-and-stick colored by atom, and the protein in gray. The three views highlight the steric clash of the N-methyl group on compound 5 with the protein. Val 163 (shown as sticks in the bottom views) is the residue that conflicts with the N-methyl substituent.
The protein residues present in the immediate vicinity of the Zn2+ ion in different MMP isoforms are generally conserved. The residues surrounding the Zn2+ ion in MMP-8 and MMP-12 are identical, which would explain the similar inhibition trends portrayed by the compounds against these MMPs. In contrast, MMP-3 has a polar Asn 162 residue in place of Gly162, Gly158, and Gly179 found in MMP-2, MMP-8, and MMP-12, respectively (Figure S1). This may contribute to the reduced potency of compound 3 (relative to compound 2) against MMP-3. The 6-methyl substituent on the pyrone ring of 3 makes the ZBG more hydrophobic relative to 2. The more polar active site environment generated by Asn162 in MMP-3 may disfavor binding of inhibitors with more hydrophobic ZBGs, hence, the difference in potency observed between compounds 2 and 3. This may also explain the observation of MMPi 1 and 3 being more potent than 2 in the hydrophobic active sites of MMP-8 and MMP-12 versus the more polar active site of MMP-3. Variation in active site polarity has been previously invoked to explain differences in the potency of a thiirane-based inhibitor on two structurally homologous zinc metalloproteinases, TACE and MMP-2.[44] Together these results highlight the importance of the ZBG and the interactions they make within the MMP active site.
Ex vivo effects of the ZBG
Perhaps the most significant effect of ZBG-modulated selectivity was observed in an ex vivo physiological model (Figure 3). Isolated perfused rat hearts were subjected to a temporary interruption of coronary flow and reflow to mimic the damage caused by IR. The isolated perfused rat heart system allows the examination of cardiac performance in the absence of influence from the nervous system or blood-borne cells.[45, 46] This system has made it possible to identify factors which can mediate IR injury such as oxidative stress, protease activation, and apoptosis.[37, 38, 47, 48] The system can also allow investigators to characterize compounds which may be able to confer cardio-protection upon an ischemic insult such as tetracyclines, antioxidants, or MMPi.[37, 48–50] It has been demonstrated that MMPs play a role in mediating ischemia-reperfusion cardiac injury and that inhibition of MMP-2 by either broad-spectrum MMPi or neutralizing antibodies significantly protects from this injury.[38] In a study by Cheung et al. use of high concentrations (100 µm) of the nonspecific MMPi o-phenanthroline or doxycycline resulted in substantial cardioprotective effects.[38] In our experimental model, 5 µm of 2 yielded essentially no improvement in the recovery of contractile function (improvements are seen at higher doses, F.J.V. unpublished results). On the other hand, 5 µm of 1 displayed an overall constant and progressive recovery after ischemia (average >80% versus controls throughout reperfusion). Finally, negative control 6, a compound that shows no substantial inhibition of MMPs, generally did not confer improvement versus the control group. As the difference in the molecular structure of the three compounds tested resides only in the ZBG, it is possible to establish for this study that the ZBG, rather than the peptidomimetic back-bone, modulates the ex vivo efficacy as indicated by the recovery of cardiac contractile function.
Possible origin of ZBG-based selectivity
Based on the observations reported here, several factors may play a role in ZBG-generated selectivity in MMPi. First, ring sterics can affect inhibitor binding. As described above, compounds 4 and 5 are poor MMPi, as addition of even a methyl group at the 1-position on the ring is not well accommodated by the MMP active sites. In contrast, substitution at the 6-position of the ring, as in inhibitor 3, is tolerated by several MMPs, and can be used as a means to select against MMP-2. This suggests a second ZBG effect that may be important, which is hydrophobicity. In general the MMP active site immediately surrounding the Zn2+ ion is relatively hydrophobic, but small differences in active site polarity can be exploited by changing the hydrophobicity and dipole of the ZBG. A third possible parameter in modulating ZBG-mediated selectivity is the pKa of the chelating ZBG. In hydroxamic acid ZBGs, as well as all of the ZBGs examined here, there is a labile hydroxy proton that likely dissociates upon Zn2+ binding. It is known that different MMP isoforms operate at different optimal pH values,[51–53] and it has been shown that the protonation state of an MMPi can have a significant impact on inhibition.[52] Therefore, the differences in pKa of different ZBGs may play a role in the inhibition and, thereby, the selectivity of a given inhibitor. The ZBGs examined in this study have pKa values that span more than three log units,[54] and studies are underway to examine the role that the pKa values might play in the inhibitory ability of these compounds. All of the aforementioned factors will have an influence on the overall, preferred binding orientation of a ZBG in the MMP active site. The overall conformation of the bound ZBG will have consequences on the strength of the Zn2+-inhibitor interaction, as well as on the positioning of the backbone toward the subsite(s). The synergism/interplay between the ZBG and backbone may also give rise to new patterns of isoform selectivity, which could not be observed by only examining hydroxamic acid based MMPi. Ongoing computational and structural studies with even more inhibitors will be required to elucidate which of the aforementioned features is most important for tuning MMPi selectivity.
Conclusions
The results presented here are the first to demonstrate that a change in ZBG can modulate, as opposed to simply abrogate, the potency and selectivity observed for MMPi against different MMP isoforms. It is proposed that the backbone substituent can be used to obtain an initial degree of selectivity (for example, shallow versus deep pocket S1′ MMPs), whereas the ZBG can then be optimized to fine tune inhibition to obtain further specificity. Furthermore, results from an ex vivo rat heart system demonstrate that the choice of ZBG can also have a substantial impact on inhibitor efficacy in a biological model. This latter finding indicates that to discover MMPi with better clinical prospects, molecular platforms other than hydroxamic acids should be explored. Recent work in the literature has suggested that ZBGs with weaker binding constants may be one route to obtaining more selective MMPi.[55] In contrast, the unique findings reported herein strongly suggest that high affinity ZBGs can be used as the basis for new patterns of selective inhibition against MMPs. Ongoing studies using in vitro, as well as relevant preclinical systems, will be required to fully exploit this new discovery and to wholly understand the origin of the observed trends in selectivity.
Experimental Section
Synthesis and characterization of MMPi
The detailed synthesis and characterization of inhibitors 1–7 are provided in the Supporting Information. Unless otherwise noted, all chemicals were purchased from commercial suppliers (Aldrich and Fisher) and used as received. 1H/13C NMR spectra were recorded at ambient temperature on a 300, 400, or 500 MHz Varian FT-NMR instruments, located in the Department of Chemistry and Biochemistry, University of California San Diego. Mass spectra were obtained at the Small Molecule Mass Spectrometry Facility in the Department of Chemistry and Biochemistry at the University of California, San Diego. Elemental Analysis was performed by NuMega Resonance Labs, San Diego.
Recombinant MMP assays
IC50 values of each MMPi against MMP-1, -2, -3, -7, -8, -9, -12, and -13 were determined using a commercially available fluorescence-based assay kit as previously described.[31, 33] All experiments were repeated in quadruplicate.
Enzyme kinetic assays
Kinetic measurements were performed using the same fluorescence-based assay kit as above to determine Vmax and Km. Ki was calculated from the Michaelis–Menten equation below:[41]
| (1) |
where [I]=inhibitor concentration at the IC50 value and [S]=initial substrate concentration used to determine IC50 value.
| (2) |
To generate the Dixon plot, initial velocities for the enzyme at different concentrations of substrate were calculated from the Michaelis–Menten equation below.[41] The x-coordinate value corresponding to the intersection of the lines for different substrate concentrations is equal to the −Ki.
Computational modeling studies
Computational modeling studies were performed on a PC workstation using a Linux (Red Hat) operating system. Figure 5 uses a published, minimized computational model of MMPi 3 in the active site of MMP-3. The model for MMPi 5 was created by simply modifying the ZBG using Insight II (using standard bond lengths and angles provided by the program) starting with the structure of 3, without further minimization or optimization of the position of the amide linker or biphenyl backbone.[32] Hence, the images for MMPi 5 do not possess novel binding interactions in the S1′ pocket. Superpositions of the various ZBGs in the MMP active sites were performed as previously reported (Figure S1).[56, 57] PDB codes for the MMP structures are: MMP-2 (1QIB),[58] MMP-3 (1G4K),[59] MMP-8 (1ZP5),[60] and MMP-12 (1RMZ).[61]
Heart preparation and perfusion
Animal studies were performed according to guidelines by the American Association for Accreditation of Laboratory Animal Care, and protocols were approved by the University of California San Diego Institutional Animal Care and Use Committee. Male Sprague–Dawley rats (250–300 g) were used for the experiments. Hearts were rapidly excised from ketamine-anesthetized animals and briefly rinsed by immersion in ice-cold Krebs–Henseleit (KH) solution. They were retrogradely perfused via the aorta at a constant pressure of 60 mmHg with non-recirculating KH buffer at 37 °C. The composition of the buffer was (mm): NaCl (118), KCl (4.7), KH2PO4 (1.2), MgSO4 (1.2), CaCl2 (2.5–3.0), NaHCO3 (25), glucose (11), and EDTA (0.5). The buffer was continuously gassed with 95% O2/5% CO2 (pH 7.4). Spontaneously beating hearts were used in all experiments. A water-filled cling-wrap balloon connected to a pressure transducer was inserted into the left ventricle through an incision in the left atrium and through the mitral valve and the volume was adjusted to achieve an end diastolic pressure of 8–12 mm Hg. A water-jacketed glass chamber was positioned around the heart to maintain its temperature at 37°C. Stock solutions of the various MMPi were initially solubilized in DMSO and subsequently diluted at the desired MMPi concentrations with KH buffer. The MMPi were provided to the isolated beating hearts both during the stabilization and reperfusion periods (i.e. throughout the experiment). Final concentrations used of DMSO in KH buffer were <0.005 %. Control hearts were perfused with the KH buffer with a comparable concentration of DMSO. The time interval between thoracotomy and attachment of the heart to the perfusion system for initiation of the stabilization period was <1 min.
Ischemia and reperfusion protocol
After 10 min of aerobic perfusion to achieve steady-state conditions, hearts were subjected to 20–25 min of global no-flow ischemia induced by clamping of the aortic inflow line. This was followed by 30 min of aerobic reperfusion as the clamp was removed. Using this IR protocol hearts developed “stunning” conditions during reperfusion. Stunning is defined by the recovery of contractile function in hearts to ~50% of that observed during the stabilization period.
Data analysis
Heart rate and left ventricular pressure were monitored on a polygraph and recorded on a computer system for subsequent analysis. Left ventricular developed pressure (LVDP) was calculated as the difference between peak systolic and end diastolic pressures of the left ventricular pressure trace. The work index, rate-pressure product (RPP), was calculated as the product of heart rate and left ventricular developed pressure. Data are expressed as mean ±SEM. Student’s t-test was used to determine the statistical significance of the data. A value of P<0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
The authors would like to thank Faith E. Jacobsen for assistance with the computational modeling studies, Dr. Cesar Oliveira for preparation of Figure 5, and Dr. Yongxuan Su for obtaining the mass spectrometry data. This work has been supported by grants from the National Institutes of Health (HL080049-01, SMC and FJV; HL-43617, FJV), the American Heart Association (0430009N, SMC), and a CONACYT-UC MEXUS doctoral fellowship to DRP. Seth M. Cohen is a Cottrell Scholar of the Research Corporation.
Footnotes
Supporting information for this article is available on the WWW under http://www.chemmedchem.org or from the author.
References
- 1.Nagase H, Woessner JF., Jr J. Biol. Chem. 1999;274:21491. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 2.Murphy GJP, Murphy G, Reynolds JJ. FEBS Lett. 1991;289:4. doi: 10.1016/0014-5793(91)80895-a. [DOI] [PubMed] [Google Scholar]
- 3.Page-McCaw A, Ewald AJ, Werb Z. Nat. Rev. Mol. Cell Biol. 2007;8:221. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skiles JW, Gonnella NC, Jeng AY. Curr. Med. Chem. 2004;11:2911. doi: 10.2174/0929867043364018. [DOI] [PubMed] [Google Scholar]
- 5.Nelson AR, Fingleton B, Rothenberg ML, Matrisian LM. J. Clin. Oncol. 2000;18:1135. doi: 10.1200/JCO.2000.18.5.1135. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker MF, Floyd CD, Brown P, Gearing AJH. Chem. Rev. 1999;99:2735. doi: 10.1021/cr0100345. [DOI] [PubMed] [Google Scholar]
- 7.Creemers EEJM, Cleutjens JPM, Smits JFM, Daemen MJAP. Circ. Res. 2001;89:201. doi: 10.1161/hh1501.094396. [DOI] [PubMed] [Google Scholar]
- 8.Lindsey ML. Heart Failure Rev. 2004;9:7. doi: 10.1023/B:HREV.0000011390.44039.b7. [DOI] [PubMed] [Google Scholar]
- 9.Reynolds JJ, Hembry RM, Meikle MC. Adv. Dent. Res. 1994;8:312. doi: 10.1177/08959374940080022701. [DOI] [PubMed] [Google Scholar]
- 10.Maeda A, Sobel RA. J. Neuropathol. Exp. Neurol. 1996;55:300. doi: 10.1097/00005072-199603000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Basset P, Bellocq JP, Wolf C, Stoll I, Hutin P, Limacher JM, Podhajcer OL, Chenard MP, Rio MC, Chambon P. Nature. 1990;348:699. doi: 10.1038/348699a0. [DOI] [PubMed] [Google Scholar]
- 12.Coussens LM, Fingleton B, Matrisian LM. Science. 2002;295:2387. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 13.Beckett RP, Whittaker M. Expert Opin. Ther. Pat. 1998;8:259. [Google Scholar]
- 14.Jacobsen FE, Lewis JA, Cohen SM. ChemMedChem. 2007;2:152. doi: 10.1002/cmdc.200600204. [DOI] [PubMed] [Google Scholar]
- 15.Overall CM, Kleifeld O. Br. J. Cancer. 2006;94:941. doi: 10.1038/sj.bjc.6603043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher JF, Mobashery S. Cancer Metastasis Rev. 2006;25:115. doi: 10.1007/s10555-006-7894-9. [DOI] [PubMed] [Google Scholar]
- 17.Lukacova V, Zhang Y, Mackov M, Baricic P, Raha S, Calvo JA, Balaz S. J. Biol. Chem. 2004;279:14194. doi: 10.1074/jbc.M313474200. [DOI] [PubMed] [Google Scholar]
- 18.Puerta DT, Cohen SM. Curr. Top. Med. Chem. 2004;4:1551. doi: 10.2174/1568026043387368. [DOI] [PubMed] [Google Scholar]
- 19.Rush TS, III, Powers R. Curr. Top. Med. Chem. 2004;4:1311. doi: 10.2174/1568026043387999. [DOI] [PubMed] [Google Scholar]
- 20.Verma RP, Hansch C. Bioorg. Med. Chem. 2007;15:2223. doi: 10.1016/j.bmc.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Puerta DT, Cohen SM. Inorg. Chem. 2002;41:5075. doi: 10.1021/ic0204272. [DOI] [PubMed] [Google Scholar]
- 22.Hajduk PJ, Shuker SB, Nettesheim DG, Craig R, Augeri DJ, Betebenner D, Albert DH, Guo Y, Meadows RP, Xu L, Michaelides M, Davidsen SK, Fesik SW. J. Med. Chem. 2002;45:5628. doi: 10.1021/jm020160g. [DOI] [PubMed] [Google Scholar]
- 23.Rao BG. Curr. Pharm. Des. 2005;11:295. doi: 10.2174/1381612053382115. [DOI] [PubMed] [Google Scholar]
- 24.Cuniasse P, Devel L, Makaritis A, Beau F, Georgiadis D, Matziari M, Yiotakis A, Dive V. Biochimie. 2005;87:393. doi: 10.1016/j.biochi.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 25.Park HI, Jin Y, Hurst DR, Monroe CA, Lee S, Schwartz MA, Sang Q-XA. J. Biol. Chem. 2003;278:51646. doi: 10.1074/jbc.M310109200. [DOI] [PubMed] [Google Scholar]
- 26.Skiles JW, Gonnella NC, Jeng AY. Curr. Med. Chem. 2001;8:425. doi: 10.2174/0929867013373417. [DOI] [PubMed] [Google Scholar]
- 27.O’Brien PM, Ortwine DF, Pavlovsky AG, Picard JA, Sliskovic DR, Roth BD, Dyer RD, Johnson LL, Man CF, Hallak H. J. Med. Chem. 2000;43:156. doi: 10.1021/jm9903141. [DOI] [PubMed] [Google Scholar]
- 28.Castelhano AL, Billedeau R, Dewdney N, Donnelly S, Horne S, Kurz LJ, Liak TJ, Martin R, Uppington R, Yuan Z, Krantz A. Bioorg. Med. Chem. Lett. 1995;5:1415. [Google Scholar]
- 29.Jacobsen FE, Lewis JA, Cohen SM. J. Am. Chem. Soc. 2006;128:3156. doi: 10.1021/ja057957s. [DOI] [PubMed] [Google Scholar]
- 30.Puerta DT, Cohen SM. Inorg. Chem. 2003;42:3423. doi: 10.1021/ic026029g. [DOI] [PubMed] [Google Scholar]
- 31.Puerta DT, Lewis JA, Cohen SM. J. Am. Chem. Soc. 2004;126:8388. doi: 10.1021/ja0485513. [DOI] [PubMed] [Google Scholar]
- 32.Puerta DT, Mongan J, Tran BL, McCammon JA, Cohen SM. J. Am. Chem. Soc. 2005;127:14148. doi: 10.1021/ja054558o. [DOI] [PubMed] [Google Scholar]
- 33.Puerta DT, Griffin MO, Lewis JA, Romero-Perez D, Garcia R, Villarreal FJ, Cohen SM. J. Biol. Inorg. Chem. 2006;11:131. doi: 10.1007/s00775-005-0053-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y-M, Fan X, Chakaravarty D, Xiang B, Scannevin RH, Huang Z, Ma J, Burke SL, Karnachi P, Rhodes KJ, Jackson PF. Bioorg. Med. Chem. Lett. 2007 doi: 10.1016/j.bmcl.2007.10.045. DOI:doi:10.1016/j.bmcl.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y-M, Fan X, Yang S-M, Scannevin RH, Burke SL, Rhodes KJ, Jackson PF. Bioorg. Med. Chem. Lett. 2007 doi: 10.1016/j.bmcl.2007.10.049. DOI:doi:10.1016/j.bmcl.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 36.Johnson LL, Bornemeier DA, Janowicz JA, Chen J, Pavlovsky AG, Ortwine DF. J. Biol. Chem. 1999;274:24881. doi: 10.1074/jbc.274.35.24881. [DOI] [PubMed] [Google Scholar]
- 37.Schulz R. Annu. Rev. Pharmacol. Toxicol. 2007;47:211. doi: 10.1146/annurev.pharmtox.47.120505.105230. [DOI] [PubMed] [Google Scholar]
- 38.Cheung PY, Sawicki G, Wozniak M, Wang W, Radomski MW, Schulz R. Circulation. 2000;101:1833. doi: 10.1161/01.cir.101.15.1833. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi R, Jin X, Cook GR. Bioorg. Med. Chem. Lett. 2007 doi: 10.1016/j.bmcl.2007.10.020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim NC, Schuster JV, Porto MC, Tanudra MA, Yao L, Freake HC, Bruckner C. Inorg. Chem. 2005;44:2018. doi: 10.1021/ic048905r. [DOI] [PubMed] [Google Scholar]
- 41.Segel IH. Enzyme Kinetics—Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: Wiley Classics Library; 1993. [Google Scholar]
- 42.Cortés A, Cascante M, Cárdenas ML, Cornish-Bowden A. Biochem. J. 2001;357:263. doi: 10.1042/0264-6021:3570263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanessian S, MacKay DB, Moitessier N. J. Med. Chem. 2001;44:3074. doi: 10.1021/jm010096n. [DOI] [PubMed] [Google Scholar]
- 44.Solomon A, Rosenblum G, Gonzales PE, Leonard JD, Mobashery S, Milla ME, Sagi I. J. Biol. Chem. 2004;279:31646. doi: 10.1074/jbc.M401310200. [DOI] [PubMed] [Google Scholar]
- 45.Ytrehus K. Pharmacol. Res. 2000;42:193. doi: 10.1006/phrs.2000.0669. [DOI] [PubMed] [Google Scholar]
- 46.Sutherland FJ, Hearse DJ. Pharmacol. Res. 2000;41:613. doi: 10.1006/phrs.1999.0653. [DOI] [PubMed] [Google Scholar]
- 47.Kang PM, Izumo S. Trends Mol. Med. 2003;9:177. doi: 10.1016/s1471-4914(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 48.Ferrari R, Guardigli G, Mele D, Percoco GF, Ceconi C, Curello S. Curr. Pharm. Des. 2004;10:1699. doi: 10.2174/1381612043384718. [DOI] [PubMed] [Google Scholar]
- 49.Scarabelli TM, Stephanou A, Pasini E, Gitti G, Townsend P, Lawrence K, Chen-Scarabelli C, Saravolatz L, Latchman D, Knight R, Gardin J. J. Am. Coll. Cardiol. 2004;43:865. doi: 10.1016/j.jacc.2003.09.050. [DOI] [PubMed] [Google Scholar]
- 50.Griffin MO, Jinno M, Miles LA, Villarreal FJ. Mol. Cell. Biochem. 2005;270:1. doi: 10.1007/s11010-005-2540-3. [DOI] [PubMed] [Google Scholar]
- 51.Cha J, Pedersen MV, Auld DS. Biochemistry. 1996;35:15831. doi: 10.1021/bi962085f. [DOI] [PubMed] [Google Scholar]
- 52.Johnson LL, Pavlovsky AG, Johnsoni AR, Janowicz JA, Man CF, Ortwine DF, Purchase CF, II, White AD, Hupe DJ. J. Biol. Chem. 2000;275:11026. doi: 10.1074/jbc.275.15.11026. [DOI] [PubMed] [Google Scholar]
- 53.Fasciglione GF, Marini S, D’Alessio S, Politi V, Coletta M. Biophys. J. 2000;79:2138. doi: 10.1016/S0006-3495(00)76461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorden AEV, Xu J, Raymond KN, Durbin P. Chem. Rev. 2003;103:4207. doi: 10.1021/cr990114x. [DOI] [PubMed] [Google Scholar]
- 55.Tochowicz A, Maskos K, Huber R, Oltenfreiter R, Dive V, Yiotakis A, Zanda M, Bode W, Goettig P. J. Mol. Biol. 2007;371:989. doi: 10.1016/j.jmb.2007.05.068. [DOI] [PubMed] [Google Scholar]
- 56.Puerta DT, Schames JR, Henchman RH, McCammon JA, Cohen SM. Angew. Chem. 2003;115:3902. doi: 10.1002/anie.200351433. Angew. Chem. Int. Ed.2003, 42, 3772. [DOI] [PubMed] [Google Scholar]
- 57.Jacobsen FE, Lewis JA, Heroux KJ, Cohen SM. Inorg. Chim. Acta. 2007;360:264. [Google Scholar]
- 58.Dhanaraj V, Williams MG, Ye Q-Z, Molina F, Johnson LL, Ortwine DF, Pavlovsky A, Rubin JR, Skeean RW, White AD, Humblet C, Hupe DJ, Blundell TL. Croat. Chem. Acta. 1999;72:575. [Google Scholar]
- 59.Dunten P, Kammlott U, Crowther R, Levin W, Foley LH, Wang P, Palermo R. Protein Sci. 2001;10:923. doi: 10.1110/ps.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campestre C, Agamennone M, Tortorella P, Preziuso S, Biasone A, Gavuzzo E, Pochetti G, Mazza F, Hiller O, Tschesche H, Consalvi V, Gallina C. Bioorg. Med. Chem. Lett. 2006;16:20. doi: 10.1016/j.bmcl.2005.09.057. [DOI] [PubMed] [Google Scholar]
- 61.Bertini I, Calderone V, Cosenza M, Fragai M, Lee YM, Luchinat C, Mangani S, Terni B, Turano P. Proc. Natl. Acad. Sci. USA. 2005;102:5334. doi: 10.1073/pnas.0407106102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.