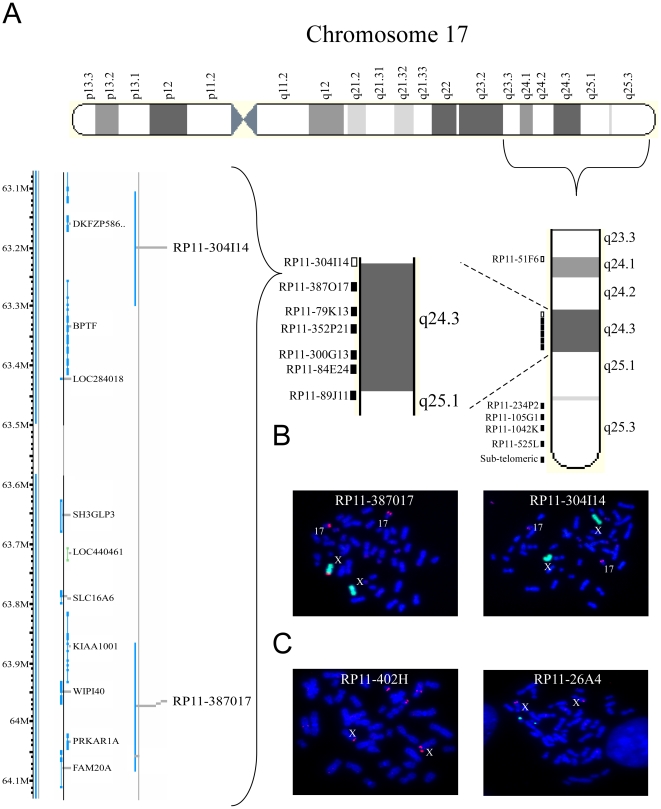
Figure 3. Mapping of the der(X)t(X;17) breakpoint.
(A) A scheme of the bacterial artificial chromosome (BAC) probes used for the FISH analysis in order to localize the translocation. Filled squares represent probes detecting 17q material on both 17q and Xq, empty squares represent probes detecting 17q material only on 17q and not on Xq. A magnification of the breakpoint region is illustrated on the left-hand side of the panel. (B) The breakpoint region is localized in the 17q24.3 region. Fluorescence in situ hybridization analysis along the 17q arm of WI-38THP-1 reveals the breakpoint region in the interval between two probes: RP11-387O17 (left-hand side – red) which is detected on chrX in addition to its normal position on chr17. In contrast, probe RP11-304I14 (right-hand side – red) is detected only on the two normal copies of chr17 and not on chrX. ChrX material is marked in green. (C) No aberrations in chromosome Xq material are detected. Fluorescence in situ hybridization along Xq arm. The two probes detecting the most distant area of the Xq arm (RP11-304H and RP11-26A - red) are visible only on the two copies of chrX and not on chr17. The chr17 centromeric region is marked in green.