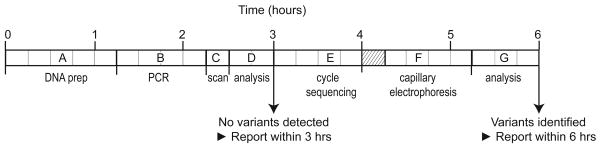
Fig 3.
Workflow diagram for CYBB mutation scanning by high resolution melting analysis. Wild type samples containing no variants can be reported within 3 hours. Rare samples with aberrant melting profiles are sequenced for variant identification and can be reported within 6 hours. (A) DNA preparation from whole blood in 60 min (Roche MagNA Pure Compact) with quantification and dilution in 15 min (NanoDrop ND-8000 Spectrophotometer), (B) PCR preparation in 15 min with PCR cycling in 45 min (BioRad C1000), (C) Mutation scanning in 15 min (Idaho Techonology LightScanner 96), (D) Scanning data analysis in 30 min → if no variants detected, report within 3 hours, (E) Cycle sequence preparation (PCR product dilution, master mix preparation, and plate loading) in 30 min followed by cycle sequencing in 45 min (BioRad C1000), (F) Capillary electrophoresis preparation (ABI 3130xl) and product clean-up in 30 min with capillary electrophoresis in 45 min, (G) Sequencing data analysis in 45 min → report variant(s) within 6 hours. The hatched area indicates overlap of cycle sequencing and capillary electrophoresis preparation.