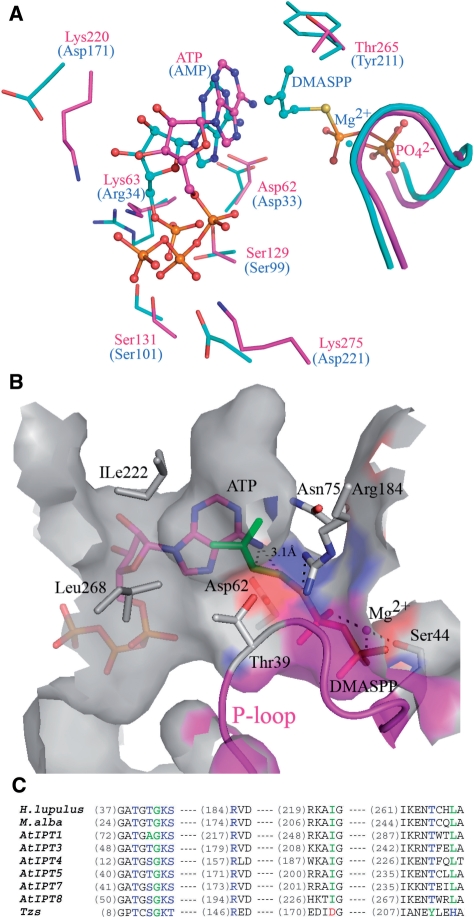
Figure 4.
Comparison of HlAIPT with Agrobacterium AIPT. (A) Superposition of adenine nucleotide and dimethylallyl group binding sites of HlAIPT (magenta) and Agrobacterium AIPT (cyan). (B) The modeling shows that the pyrophosphate group of DMAPP binds to the P-loop and is coordinated by an Mg2+ ion. Ile222 and Leu268 surrounding the hydrophobic end of the dimethylallyl group may contribute to the substrate specificity. (C) Sequence alignment of AIPTs. The amino acid residues involving prenyl-donor substrate specificity are shown in blue, green and red according to their types. AtIPTs are isozymes from A. thaliana (17). Asp173, Tyr211 and His214 of Agrobacterium AIPT (Tzs) are substituted by Ile222, Thr265 and Leu268 in HlAIPT, respectively.