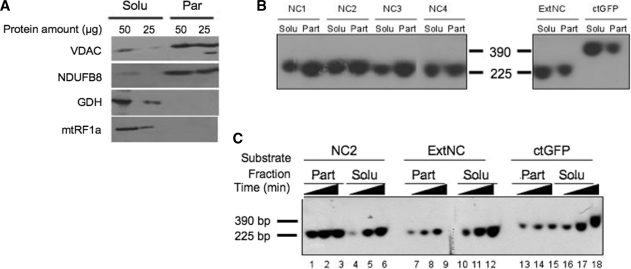
Figure 1.
Sequence-dependent association of imported DNA with the mitochondrial particulate fraction. (A) Mitochondrial sub-fractionation. Isolated rat liver mitochondria were disrupted by repeated freezing and thawing. Soluble (Solu) and particulate (Part) fractions were separated by centrifugation at 100 000g for 1 h and the indicated amount of protein from the different fractions was subjected to western blot analysis. Purity of fractions was assessed by blotting with antibodies against marker proteins. The VDAC and a component of respiratory chain complex I (NDUFB8) are outer and inner membrane markers, respectively; glutamate dehydrogenase (GDH) and the mitochondrial translation release factor (mtRF1a) are matrix soluble markers. (B) NCR corresponding sequences show a strong association with the particulate fraction after import into isolated rat mitochondria. Four probes ranging from 221 to 235 bp (NC1–NC4) spanning most of the NCR, an additional mtDNA probe from outside the NCR (ExtNC) and a 390 bp probe from the GFP coding sequence (ctGFP) were PCR amplified and internally labelled with [α32P]dCTP (primers and locations are given in Supplementary Table S1). Following a 45-min incubation with coupled rat mitochondria, non-imported DNA was DNase I digested, mitochondria sub-fractionated, protected DNA was extracted and subjected to non-denaturing gel electrophoresis prior to autoradiography and visualization as detailed in ‘Materials and Methods’ section. Typical results are shown from three repeats. Solu and Part refer to the soluble and particulate mitochondrial fractions, respectively. Markers refer to labelled probes (NC2 and ctGFP) of the indicated sizes. (C) Imported NCR DNA associates rapidly with the particulate fraction in a saturable process. A representative probe for the NCR, NC2 and the two additional probes ExtNC and ctGFP were internally labelled as in Figure 2B. Incubation of mitochondria with each probe was terminated after 15, 30 or 45 min (indicated as increasing time) before sub-fractionation, DNA extraction and visualization as for Figure 2B. Typical results are shown from three repeats.