Abstract
Plasmids, conjugative transposons and phage frequently encode anti-restriction proteins to enhance their chances of entering a new bacterial host that is highly likely to contain a Type I DNA restriction and modification (RM) system. The RM system usually destroys the invading DNA. Some of the anti-restriction proteins are DNA mimics and bind to the RM enzyme to prevent it binding to DNA. In this article, we characterize ArdB anti-restriction proteins and their close homologues, the KlcA proteins from a range of mobile genetic elements; including an ArdB encoded on a pathogenicity island from uropathogenic Escherichia coli and a KlcA from an IncP-1b plasmid, pBP136 isolated from Bordetella pertussis. We show that all the ArdB and KlcA act as anti-restriction proteins and inhibit the four main families of Type I RM systems in vivo, but fail to block the restriction endonuclease activity of the archetypal Type I RM enzyme, EcoKI, in vitro indicating that the action of ArdB is indirect and very different from that of the DNA mimics. We also present the structure determined by NMR spectroscopy of the pBP136 KlcA protein. The structure shows a novel protein fold and it is clearly not a DNA structural mimic.
INTRODUCTION
DNA restriction and modification (RM) systems are widespread in bacteria and archaea, and function as defence systems to reduce the influx of foreign DNA on mobile genetic elements via transduction, transformation and conjugation (1–3). The RM systems can be classified into four types, I–IV, depending upon their complexity of structure and function (2) with the Type I RM enzymes being the most complex exhibiting both restriction endonuclease and modification methyltransferase activities in one large complex although a sub-assembly can act as a monofunctional modification methyltransferase (4). These enzymes are composed of three subunits encoded by the genes hsdR, hsdM and hsdS (hsd denotes ‘host specificity of DNA’). HsdM (∼50–60 kDa) and HsdS (∼50 kDa) subunits comprise a methyltransferase (Mtase, M2S1), which in association with HsdR forms an endonuclease (R2M2S1). The HsdR (∼140 kDa) subunits are required for restriction, the HsdS subunit specifies the DNA target sequence and the HsdM subunits catalyses the methylation reaction. Depending upon the methylation state of the DNA, the RM complex can function as either an endonuclease (REase) or an Mtase. The REase will cleave incoming DNA that has not been appropriately modified by the Mtase. If the target sequence is unmodified (unmethylated) then it is targeted for restriction. If the target sequence is hemi-methylated, the unmethylated target strand also becomes methylated as well. Fully methylated DNA is immune to restriction. The Type I RM systems are further subdivided into families, five of which are defined at present by DNA hybridization, subunit complementation, antibody cross-reactivity and sequence conservation.
These complex Type I systems are found in over 50% of bacterial species (5) and this defence can be extremely strong (6). They therefore place a considerable evolutionary pressure on mobile genetic elements to evolve anti-restriction countermeasures encoded by the foreign DNA (2,3,7). One such measure is the use of anti-restriction proteins to modify or inactivate the RM system. These are frequently targeted against Type I RM systems and operate in a variety of different manners. The simplest anti-restriction proteins mimic the structure of DNA and include the proteins ocr from phage T7 and ArdA from plasmids and conjugative transposons (8,9). The ocr protein from phage T7 (9,10) and the ArdA proteins from conjugative plasmids and conjugative transposons (3,8,11–15) have been the most intensively studied of the DNA mimics. These inactivate the host’s Type I RM enzyme by binding in the substrate (DNA) binding groove (16,17). The structure of these proteins shows they mimic DNA both in size and surface charge distribution and have high affinity for binding to the RM enzyme. Another group are single-strand DNA binding proteins and represented by the ArdC protein (7,18). Binding to the single-stranded DNA intermediate formed during conjugation seems to lead to anti-restriction though why this occurs is unclear as the Type I RM systems act on double-stranded DNA (11,18). The ArdB proteins (7,11,18) and their close homologues the KlcA proteins (19,20) are the subject of this study.
In annotated sequence databases, the ArdB proteins appear to be mostly confined to the proteobacteria. For instance, Escherichia coli K12 W3110 contains two prophages on its chromosome, CP4-6 and CP4-57, encoding ardB homologues termed yafX and yfjX, respectively (21). Comparison of the predicted products of these two genes with sequence databases indicates that they are highly homologous to the ArdB protein from the IncN incompatibility group conjugative plasmid pKM101 (11). Similarly, another putative ardB, ORF14, is encoded by the second pathogenicity island, PAI IICFT073, from the uropathogenic E. coli CFT073 (22,23). Putative homologues have also been identified in Legionella pneumophila (24) and Achromobacter denitrificans (25). The ArdB proteins are small in size with an acidic nature, which is similar to ocr and ArdA. The anti-restriction function for ardB from pKM101 has been demonstrated but it was also observed that it did not inhibit modification (11) in contrast to the ocr and ArdA DNA mimics. This inability to block modification occurred despite the presence of the short ‘anti-restriction amino acid motif’ identified in ocr, ArdA and ArdB (7,26). The recent atomic structure of ArdA indicates that this motif has a structural role in maintaining the fold of ArdA rather than a direct role in inhibiting the RM enzyme (8). Therefore, the presence of this motif in ArdB does not necessarily imply a functional role.
klcA is one of three genes found on the kilC operon, one of four kil loci (20) found on IncP plasmids. Expression of kil genes can be lethal to E. coli and is regulated by the kor genes. korA and korC control klcA on the broad host range plasmid RK2. klcA from RK2 showed 31% identity and 56% similarity to ardB from pKM101. Plasmids of the incompatibility group P are found in a wide range of Gram-negative bacteria (27). IncP-1β members usually carry multiple antibiotic resistance determinants and genes for the degradation of xenobiotic compounds (28). An exception is the IncP-1β plasmid pA1 from Sphingomonas sp. A1 that does not contain any of the typical accessory genes of IncP-1βplasmids (29). Similarly, pBP136, isolated from Bordetella pertussis in a fatal case of whooping cough (19) was found to contain no accessory genes for antibiotic resistance, remnants of insertion sequences or transposons. It contains two regions involved in plasmid conjugation and a third region involved in plasmid replication, central control, stable inheritance and partitioning. This region contained a klcA gene encoding a protein with 71% identity to KlcA from RK2. Sequence alignment revealed that KlcA from pBP136 is also significantly related to the anti-restriction ArdB proteins (19) showing, e.g. 30% identity and 46% similarity to the 16.5 kDa ArdB from the IncN plasmid pKM101 (11). The significant homology of KlcA to ArdB implied a possible function of KlcA as an anti-restriction protein. However, klcA from the IncP-1α plasmid RK2 showed no anti-restriction activity against the archetypal Type I RM system EcoKI from E. coli (20).
Here, we demonstrate that four ardB and two klcA display an anti-restriction function in vivo against Type I RM systems but contrary to expectations, we find that in vitro, the purified ArdB proteins are unable to prevent the normal restriction endonuclease activity of the EcoKI Type I RM enzyme indicating an anti-restriction mechanism distinct from the DNA mimicry used by the ArdA anti-restriction proteins (8,12,13). We present the NMR-derived atomic resolution structure of the full length, 142 amino acid, KlcA protein from plasmid pBP136 isolated from B. pertussis and show that it has a novel fold and clearly is not a DNA structural mimic.
METHODS
Strains, plasmids and genes
Escherichia coli JM109 was used as a general cloning strain (Promega Madison, WI, USA). The expression strain E. coli BL21(DE3) was purchased from Invitrogen (Groningen, The Netherlands). The E. coli strains used for assessing restriction activity in vivo comprise NM1261 (r−m−), NM1049 (restriction system Type IA), NK354 (Type IB), NK402 (Type IC), NM1009 (Type ID) and were a kind gift of Professor Noreen E. Murray (School of Biology, University of Edinburgh, UK). NM1041 (hsd+ clpX−) was again obtained from Noreen Murray (30). The NK and NM series were converted to DE3 lysogens as described previously (8).
Four ArdB genes were identified in the public sequence database held by the National Center for Biotechnology Information (Bethesda, MD, USA), Table 1. The gene sequences were then synthesised by GeneArt (Regensburg, Germany). The codon usage pattern was optimized in order to maximize expression yield in E. coli. To facilitate cloning, an NdeI restriction site was engineered to overlap the ATG start codon of each synthetic gene. In addition, a unique HindIII site was also included just downstream of the stop codon. The synthesised genes were then ligated into pET24a (kanamycin resistant) at the NdeI and HindIII sites of the expression vector and the anticipated sequence was verified by DNA sequencing on both strands. Two further ardB genes from E. coli DH5α (yjfX and yafX), Table 1, were PCR-amplified from genomic DNA and ligated into pET20b (Novagen, Madison, WI, USA). The primers used to amplify yfjX were: forward primer, 5′-ACGGTGTCATATGACAACACAGACACAGT-3′; reverse primer, 5′-GCTAGAATTCTTAATCAATAATGCGCAT-3′. The primers used to amplify yafX were: forward primer 5′-ACGGTGTCATATGACAACACAGACGCAGC-3′; reverse primer, 5′-GCTAGAATTCTCAGTCGATAATACGCAG-3′. The NdeI and EcoRI recognition sites used in the subsequent ligation are underlined. In both cases an amplified product of the expected size was obtained. The PCR products were then ligated into the NdeI and EcoRI sites of pET20b (ampicillin resistant). The sequence of the insert was verified to ensure no mistakes had been introduced during amplification. For assessing the effect of expressing klcA136 in NM1041(DE3) (hsd+ clpX−) on growth in the presence and absence of 2-AP, klcA136 was ligated into an expression vector with a carbenicillin selection marker (pET20b). This was achieved by digesting the KlcA136 pET24a expression construct with NdeI and HindIII and ligating the insert into the corresponding sites of pET20b to produce pET20b-klcA136.
Table 1.
The source of the ardB and klcA genes investigated
| Identifier in NCBI protein database | Gene | Protein annotation | Our name | Organism | Amino acid length | Molecular weight including N-terminal methionine | Molar extinction coefficient M−1 cm−1 | Predicted isoelectric point | Optimal protein expression temperature (°C) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| NP_752219 | ORF14 | ArdB | ArdBCFT | E. coli CFT073 | 157 | 17 832.1 | 34 410 | 4.83 | 37 | (23) |
| ACB01415 | yafX | ArdB | ArdBYAF | E. coli K12/CP4-6 prophage | 152 | 17 419.5 | 37 410 | 4.76 | 37 | (21) |
| AAC75691 | yfjX | ArdB | ArdBYFJ | E. coli K12/CP4-57 prophage | 152 | 17 303.3 | 37 410 | 4.64 | 37 | (21) |
| YP_122202 | plpp0047 | ArdB | ArdBLPP | L. pneumophila str. Paris | 134 | 15 409.5 | 26 740 | 5.05 | 25 | (24) |
| BAF33451 | KlcA | KlcA | KlcA136 | B. pertussis BP136 | 142 | 15 659.6 | 22 900 | 4.84 | 30 | (19) |
| NP_990889 | pEST4011_14 | KlcA | KlcAADE | A. denitrificans | 144 | 16 026.8 | 21 620 | 4.74 | 30 | (25) |
The extinction coefficients of the proteins were calculated using the method of Gill and von Hippel (31), Table 1. All concentrations quoted for the proteins are based on the concentrations of monomers. The monomer molecular weights of the proteins are shown in Table 1 and include the N-terminal methionine.
Expression and purification of proteins
Escherichia coli BL21(DE3) cells were transformed with the recombinant plasmid and transformants selected using LB agar supplemented with 25 μg/ml kanamycin or 100 µg/ml carbenicillin. A single colony was picked and grown in LB medium plus 25 μg/ml kanamycin or 100 µg/ml carbenicillin at 37°C to an optical density at 600 nm (OD600) of 0.5. Heterologous gene expression was induced by the addition of IPTG to 1 mM and the growth continued for a further 3 h at the temperature given in Table 1 before harvesting. For four of the chosen genes, a prominent protein band corresponding to the predicted protein mass was visible on SDS–polyacrylamide gels of induced cells of BL21(DE3) containing the recombinant plasmid that was not visible in the negative control of the vector alone in BL21(DE3). The A. denitrificans klcA and L. pneumophila ardB could not be expressed at levels sufficient for purification. For protein production, a single colony of was used to inoculate 100 ml of LB medium supplemented with 25 μg/ml kanamycin or 100 µg/ml carbenicillin. The starter culture was grown to an OD600 of 0.5–0.8 at 37°C and used to inoculate 10 L of LB containing 25 μg/ml kanamycin or 100 µg/ml carbenicillin and the cells grown at 37°C to an OD600 of 0.5. Heterologous gene expression was induced as described above. Typically 50 g of cells were obtained from 10 l of culture. Cells were harvested by centrifugation (6000g, 4°C, 15 min) and the cell pellet was stored at −20°C until required.
Approximately 20 g of cell pellet was defrosted on ice for 30 min and resuspended in 200 ml buffer A (20 mM Tris–HCl pH 8.0, 7 mM 2-mercaptoethanol). A cocktail of protease inhibitors (Complete Protease Inhibitor Cocktail Tablet; Roche) was added to the cell suspension (1 tablet per 50 ml solution). The cells were disrupted by sonication on ice using a Soniprep 150 sonicator (Sanyo, Tokyo, Japan) fitted with a 9 mm diameter probe (5 × 1 min bursts with 15 s interval between each burst). The cell lysate was centrifuged at 20 000g for 1 h at 4°C and the supernatant was filtered through a 0.45 μm filter unit (Sartorius AG, Goettingen, Germany). The clarified extract was then loaded onto a 30 cm × 1 cm diameter DEAE anion exchange column equilibrated with buffer A, at a flow rate of 60 ml/h. The column was washed with ∼3 column volumes of buffer A to remove unbound material. Bound proteins were then eluted using a 500 ml gradient of 0–0.7 M NaCl in buffer A at a flow rate of 20 ml/h. Fractions that gave a UV absorbance were subsequently analysed by SDS–PAGE and those containing ArdB were pooled and concentrated using a spin concentrator with a 5 kDa MWCO (VivaScience AG, Hannover, Germany) to a final volume of ∼5 ml. The sample was then loaded on a Superdex-200 column (GE Healthcare, Uppsala, Sweden), equilibrated with buffer A + 0.2 M NaCl, and the protein was eluted at a flow rate of 10 ml/h. The fractions were subsequently analysed by SDS–PAGE and those containing ArdB were pooled and concentrated. Purified ArdB/KlcA was stored at −20°C in buffer A supplemented with glycerol to 50% (v/v).
Analysis of proteins
The methods employed for assaying ardB activity in vivo used the efficiency of plating (eop) of phage lambda (virulent), λv, on the various strains of E. coli as described previously in detail (8,13). The NM and NK E. coli strains were transformed with the derivatives of pET24a or pET20b described above but protein overexpression was not induced. Instead, we relied upon the small amount of leaky expression to produce the anti-restriction proteins in sufficient quantity to demonstrate anti-restriction in vivo. All assays were performed in triplicate and at least 50 phage plaques per plate were counted. The standard deviation on the anti-restriction and anti-modification values is ± 25%.
To assess the possible role of the ClpXP in the anti-restriction behaviour of ArdB and KlcA, the ability of E. coli NM1041 (hsd+ clpX−) transformed with plasmids expressing ardB or klcA to survive on an agar plate containing 2-aminopurine (2-AP) was assessed (32). The strain will not grow in the presence of 2-AP unless the EcoKI RM system is inactivated by the production of active ArdB or KlcA. A single colony from an agar/carbenicillin plate of E. coli NM1041 transformed with either pET20b or pET20b-klcA136 was streaked on an agar plate with or without 2-AP and incubated overnight at 37°C. This was repeated in triplicate with different colonies.
Fourier transform ion cyclotron mass spectrometry (33), isothermal titration calorimetry (10,13,33), size exclusion chromatography (34) and assays of EcoKI endonuclease activity (8,13,34) were performed as described previously. To investigate whether ArdB or KlcA interfered with the assembly of the EcoKI nuclease, 42 nM EcoKI Mtase and ∼100 nM HsdR proteins (34) were incubated separately with a 20-fold excess of ArdB or KlcA in the nuclease reaction buffer for 2 min at 37°C. Equal volumes of these two samples were then mixed and pBRsk1 DNA added to 3 nM. After incubation of this reaction mix for 8 min at 37°C, the reaction was stopped as above.
NMR spectroscopy and structure calculations
The KlcA protein from B. pertussis was uniformly labelled with 15N and 13C by growing the recombinant strain in M9-minimal medium containing 15NH4SO4 and 13C-d-glucose as the sole nitrogen and carbon sources, respectively. The labelled protein was purified as described above. Samples for NMR measurements contained 0.7 mM KlcA, 90% H2O/10% D2O and 0.05% NaN3 in 20 mM deuterated sodium acetate buffer (pH 6.0).
All NMR experiments were carried out at 298 K unless stated otherwise on a Bruker AVANCE 800 MHz with a 5-mm triple resonance cryoprobe. NMR experiments for 1H, 15N and 13C backbone and side-chain resonance assignments included: two-dimensional (2D) 1H-1H TOCSY, 1H-1H NOESY, 1H-15N HSQC, 1H-13C HSQC, (HB)CB(CGCD)HD/(HB)CB(CGCDCE)HE, three-dimensional (3D) HNCO, HN(CA)CO, CBCANH, CBCA(CO)NH, HBHA(CO)NH, HBHANH, (H)CCH-TOCSY, H(CCO)NH, C(CO)NH (35). 3D 15N- and 13C-edited NOESY-HSQC spectra were acquired to obtain distance constraints (36,37). Aromatic 1Hδ and 1Hε frequencies were obtained from (HB)CB(CGCD)HD/ (HB)CB(CGCDCE)HE spectra (38) where the aromatic 1H frequencies are correlated with the assigned 13Cβ frequencies of the aromatic residue.
NMR data were processed using the Azara suite of programs (Wayne Boucher, Department of Biochemistry, University of Cambridge, UK, http://www.ccpn.ac.uk/azara/) and analysed using Ccpnmr Analysis (39). NOE assignments were generated during the initial rounds of structure analysis using CANDID (40) in CYANA 2.1 (41). Dihedral constraints were generated using TALOS (42). The NOE assignments obtained by CANDID were checked manually and from the 100 structures calculated, 20 structures with lowest target function were selected for further analysis. The structures were further refined with the incorporation of residual dipolar couplings (RDCs) using CNS (43). PROCHECK-NMR (44) and WHATIF (45) were used to analyse the quality of the structures. MOLMOL (46) and PYMOL (http://www.pymol.org) were used for visualisation.
A 200 µM 15N-labelled KlcA sample in 20 mM deuterated sodium acetate, 0.05% NaN3, pH 6.0 in 90% H2O/10% D2O aligned with 6.25 mg/ml of Pf1 filamentous phage (Pf1 protease and RNase free, Profos) was used to measure RDCs. Magnetic alignment was monitored by 1D 2H NMR spectra as a function of phage concentration. A 7 Hz residual quadrapolar splitting of the deuterium signal was achieved with 6.25 mg/ml phage. 1H-15N splittings were measured under isotropic and partially aligned conditions using 2D IPAP 1H-15N HSQC experiments (47). Analysis and back calculation of the RDCs were performed with REDCAT (48).
Relaxation experiments were performed on a 600 MHz Bruker Avance spectrometer at 25°C with 200 µM 15N labelled KlcA. For the longitudinal (T1) and transverse (T2) relaxation data for the backbone 15N nuclei of KlcA, water was kept along the z-axis during the entire pulse sequence (49) and extra delays were used in the T1 experiment to eliminate differential heating arising from the variations of the relaxation delay (50). Eight interleaved spectra were recorded for each relaxation delay T1 (51 twice, 401, 601, 801, 901, 1001 and 1201 ms) and T2 (16 twice, 48, 64, 80, 96, 112 and 144 ms). T1 and T2 cross-peak intensities were measured from peak heights and fit to an exponential decay function I(t) = I(0)e−Rt where I(0) and I(t) are the cross-peak intensity at time 0 and time t, where time is the relaxation delay and R (1/T) is the relaxation rate (39). For the 2D 1H-15N heteronuclear NOE experiment (49) a 3 s pre-saturation period and a 2 s relaxation delay was used; the control experiment had an equivalent 5 s delay. Spectra from the heteronuclear NOE experiment were extended by linear prediction and zero filling prior to apodisation with a 5% shifted mixed Gaussian/exponential function. NOE peak heights were taken from the Gaussian fits of the frequency-domain data. The NOE was calculated as NOE = Ion/Ioff where Ion and Ioff are the peak heights with and without 1H pre-saturation, respectively. T1, T2 and NOE data were fitted using ‘Quadric_diffusion’, (AG Palmer, Columbia University, NY, USA, http://biochemistry.hs.columbia.edu/labs/palmer/software/quadric.html) to obtain the rotational diffusion tensor. Model-free analysis of the relaxation data was performed using Tensor2 (51). For this analysis a subset of relaxation data was selected using criteria defined by Tjandra et al. (52). The relative moments of inertia were calculated with ‘pdbinertia’ (http://biochemistry.hs.columbia.edu/labs/palmer/software/pdbinertia.html) and the average rotational correlation time (τc) calculated with r2r1_tm (http://biochemistry.hs.columbia.edu/labs/palmer/software/r2r1_tm.html).
The complete set of coordinates for the structure has been deposited at the Protein Data Bank (PDB) (53) with accession code 2 kmg and in the Biological magnetic resonance bank with code 16428.
Bioinformatics analysis of sequence and structure
Secondary structure was identified using STRIDE (54). Fold topology analysis was conducted using HERA (55) under PROMOTIF v2.0 (56) and schematically redrawn using TopDraw (57). The B. pertussis KlcA sequence was subjected to a BLAST search (58) against the PDB and to fold recognition server PHYRE (59) to check if there was any clear or remote homology to an existing structure or fold at the sequence level. Additionally, the 3D-coordinates of the KlcA structure were compared against 3D-fold and structure libraries using DALI (60) and SSM (61).
Sequences related to B. pertussis KlcA (UniProt Accession no: Q08L07) were obtained using a BLASTP search (58) against the UniProt sequence database (62), via the NPS@ server (63). A total of 182 sequences with an E-value of less than 10−6 were obtained and this list was made non-redundant initially by removing 62 identical sequences. The remaining 120 KlcA-like sequences were retrieved and a multiple sequence alignment was generated using the program PROMALS3D (64). This set of sequences was rendered further non-redundant by removing all 90% pairwise identical sequences in the alignment using the Expasy server (http://www.expasy.ch/tools/redundancy/). A total of 44 non-redundant divergent sequences were thus obtained. Upon analysis, one sequence that lacked the C-terminal helix 5 was discarded and three other sequences, which possessed N-terminal extensions, were trimmed. The remaining 43-aligned sequences were used as input for Consurf (65) (http://consurf.tau.ac.il/) to map sequence conservation on the surface of the protein. The electrostatic surface potential representation was calculated using GRASP (66). The lipophilic surface rendition was generated using MOLCAD (67) under SYBYL (Tripos Associates, St Louis, MO, USA). Solvent-accessibility calculations were performed using GETAREA (68).
RESULTS
The ArdB proteins from the E. coli pathogenicity island, the CP4-6 prophage, the CP4-57 prophage and L. pneumophila will be referred to as ArdBCFT, ArdBYAF, ArdBYFJ and ArdBLPP, respectively. The KlcA proteins from B. pertussis and A. denitrificans will be referred to as KlcA136 and KlcAADE, respectively. The names ArdB and KlcA without a subscript are to be understood as referring to all of the ArdB or KlcA studied.
Overexpression of klcA is not lethal to E. coli
It has been shown that unregulated expression of the kil genes leads to killing of the host cells. The cells become elongated with distorted outer membranes and macromolecular synthesis ceases (69). Previous studies of the kilC operon in plasmid RK2 showed that klcA was responsible for the lethality of E. coli host cells in the absence of korA and korC (20). However, we were able to overexpress the klcA136 gene in a pET vector in the absence of kor determinants with no apparent harmful effects. Furthermore, in vivo anti-restriction experiments performed on the klcA from RK2 failed to identify any signs of anti-restriction function (20), whereas the klcA136 and klcAADE showed efficient anti-restriction activity against all four families of Type I restriction systems as described below.
In vivo activity of ardB and klcA genes against the four families of Type I RM systems
The activity of anti-restriction genes can be tested by determining how many phages successfully infect a bacterial lawn in the presence or absence of host RM systems when the bacterium also contains a plasmid encoding the anti-restriction protein. All of the ardB and klcA genes except ardBLPP showed significant levels of anti-restriction (>10-fold enhancement in the number of phage infections) despite their low levels of expression in the absence of induction, Table 2. The poor activity of the ardBLPP and klcAADE genes were most likely due to poor expression as neither could be observed on SDS–PAGE. The effectiveness of the other proteins in inhibiting restriction matched levels observed for the ocr (9,10,15,70) and ArdA proteins (8,12,13) in vivo under similar conditions.
Table 2.
Effect of expression of six different ardB and klcA on the RM of bacteriophage λv by four different Type I RM systems, each representing one of the four families (IA–ID) of Type I RM systems
| Gene | Chromosomal restriction system in host E. coli |
|||||||
|---|---|---|---|---|---|---|---|---|
| NM1049 Type IA EcoKI |
NK354 Type IB EcoAI |
NK402 Type IC EcoR124I |
NM1009 Type ID StySBLI |
|||||
| Anti-restriction | Anti-modification | Anti-restriction | Anti-modification | Anti-restriction | Anti-modification | Anti-restriction | Anti-modification | |
| ardBCFT | 710 | 2.0 | 54 | 1.8 | 199 | 1.3 | 1174 | 6.0 |
| ardBYAF | 9250 | 1.4 | 27 | 1.6 | 7510 | 10.6 | 1080 | 1.5 |
| ardBYFJ | 6160 | 1.2 | 10 | 1.1 | 251 | 1.0 | 1650 | 18.6 |
| klcAADE | 1140 | 1.6 | 14 | 1.2 | 13 | 1.9 | 37 | 1.2 |
| klcA136 | 2430 | 1.4 | 76 | 1.1 | 628 | 3.1 | 969 | 1.4 |
| ardBLPP | 270 | 1.1 | 17 | 1.1 | 1.9 | 1.4 | 132 | 1.4 |
| Vector alone | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 |
The host E. coli contained a chromosomal copy of the RM system and an uninduced expression plasmid expressing ardB or klcA at low level. An anti-restriction or anti-modification value >2 was taken to indicate anti-restriction activity in vivo. Errors in the anti-restriction and anti-modification values are ± 25%.
However, in contrast to ocr and ArdA, anti-modification by ArdB was generally very weak (<3-fold and negligible given the limitations of the assay), Table 2. In other words, the phages that escaped restriction were easily modified by the methylation activity of the RM system and ArdB had little if any inhibitory effect on methylation. In three cases, some inhibition of methylation was observed; ArdBCFT and ArdBYFJ slightly inhibited methylation by the Type ID RM system and ArdBYAF slightly inhibited methylation by the Type IC RM system. These results show a clear difference between the activities of ArdB and KlcA compared to the DNA mimics, ocr and ArdA, suggesting that they use different mechanisms for interfering with the Type I RM systems.
KlcA136 anti-restriction function does not involve the ClpXP protease
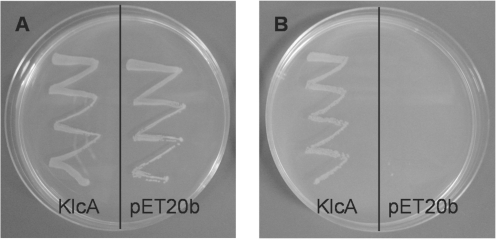
It has been shown that ClpXP-dependent regulation of the EcoKI endonuclease activity enables bacteria that acquire unmodified chromosomal target sequences to survive. When cells are under stress after prolonged exposure to treatments that damage DNA, such as exposure to 2-AP, the ClpXP-dependent pathway operates to prevent EcoKI from cleaving the chromosome (71). The restriction subunit of the nuclease (HsdR) is degraded in the presence of ClpXP (71). As described later, we did not observe any direct interaction of KlcA136 with the EcoKI in vitro; therefore we tested the hypothesis that the anti-restriction activity of KlcA136 was dependent on the ClpXP pathway. A clpX− mutant, NM1041(DE3), was plated in the presence and absence of 80 µg/ml 2-AP, Figure 1. The clpX− mutant was also transformed with the plasmid, pET20b-klcA136, expressing klcA136. As expected, the clpX− strain with no anti-restriction determinant present, grew well in the absence of 2-AP, but no growth was observed in the presence of 2-AP. In contrast, the clpX− strain carrying klcA136 grew equally well in both the presence and absence of 2-AP. This finding indicates that KlcA136 is able to block EcoKI restriction without involving the ClpXP protease. We assume that this result holds for ArdB and KlcA in general.
Figure 1.
The effect of expressing KlcA136 in a 2-AP sensitive strain of E. coli. A single colony of the ClpX− strain NM1041(DE3) harbouring either the KlcA expression construct (pET20b-klcA136) or the corresponding vector alone (pET20b) was picked with a loop and spread onto an LB agar plate supplemented with 100 μg/ml carbenicillin, 25 μg/ml kanamycin in the absence (A) or presence (B) of 2-AP (80 μg/ml). The plates were then incubated at 37°C for 14 h.
Solution state behaviour of ArdB and KlcA proteins
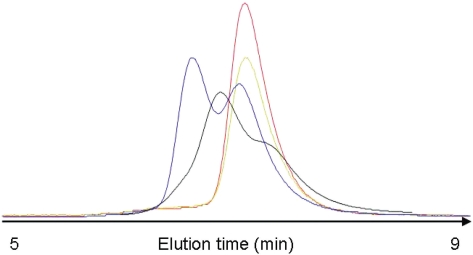
During purification it was noted that ArdBYAF and ArdBCFT behaved anomalously on a large volume (60 cm × 1.6 cm) Superdex 200 size-exclusion chromatography column despite showing only a single homogeneous species of the correct molecular weight on SDS–PAGE (data not shown) and mass spectrometry, Supplementary Table S1. Analysis with a small volume analytical column (25 cm × 0.46 cm) revealed two peaks corresponding to a dimer and a monomer for ArdBYAF while ArdBCFT showed two peaks on the large volume column and a broad peak with shoulders on the small volume column, Figure 2. ArdBYFJ and KlcA136 showed only a single monomer peak. These results indicate that there was genuine heterogeneity in the quaternary structure for two of the four proteins and the dependence on column volume for the ArdBCFT protein further suggested that the apparent switching between the monomer and dimer forms was occurring on the minute timescale of the chromatography process.
Figure 2.
An overlay of elution profiles for different proteins from a small volume (25 cm × 0.46 cm diameter) size exclusion chromatography column showing the variable quaternary structure of the four different proteins in 20 mM Tris, 20 mM MES, 0.2 M NaCl, 0.1 mM EDTA, 7 mM β-mercaptoethanol, pH 6.5: ArdBCFT (black, 4 µM)), ArdBYAF (blue, 3 µM), ArdBYFJ (red, 5 µM) and KlcA136 (green, 4 µM). The proteins were detected using tryptophan fluorescence emission and are arbitrarily scaled vertically.
In vitro activity of ArdB and KlcA136 proteins against the EcoKI Type IA restriction endonuclease
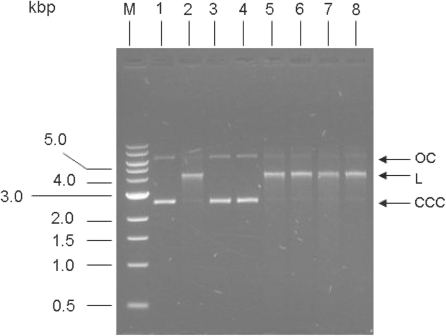
A circular plasmid containing a single copy of the target sequence for the EcoKI type I RM enzyme is efficiently cleaved by stoichiometric amounts of EcoKI in the presence of the essential cofactors SAM, ATP and Mg2+ resulting in a linearised plasmid (34). Stoichiometric amounts of the ocr and ArdA anti-restriction proteins inhibit this activity by binding strongly to and blocking the DNA binding site on EcoKI (9,12,13). Figure 3 shows that addition of a 20-fold excess of ocr or ArdA, expected to ensure complete inhibition, did indeed prevent any nuclease activity of the EcoKI restriction enzyme. Addition of the same concentration and degree of excess of any of the examined ArdB or KlcA136 proteins to the endonuclease reaction showed no observable inhibition of the nuclease activity of EcoKI. Thus the anti-restriction activity clearly observed in vivo was not due to an anti-nuclease activity in contrast to what one might expect from the results of the anti-restriction assay. Hence, none of the ArdB or KlcA136 tested appeared able to act as an inhibitor of the Type IA RM enzyme EcoKI in DNA cleavage. The order of addition of the reactants can be important for analysing Type I endonuclease activity. For instance ocr is unable to inhibit EcoKI if it is added after the DNA and ATP (72) so for this reason we added the DNA last to the mixture containing EcoKI, cofactors and anti-restriction protein to avoid this problem. Even with this precaution, ArdB and KlcA136 were unable to influence the endonuclease activity of EcoKI.
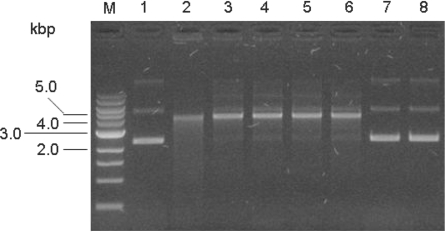
Figure 3.
Nuclease assay to test for inhibition of EcoKI by ArdB proteins. Lane 1, undigested DNA; lane 2, EcoKI digested DNA; lane 3, plus ocr; lane 4, plus Orf18 ArdA; lane 5, plus ArdBYAF; lane 6, plus ArdBYFJ; lane 7, plus KlcA136; lane 8, plus ArdBCFT; lane M, DNA ladder with sizes in kb. Plasmid DNA used was non-methylated pBRsk1. Ratio of the anti-restriction proteins (monomers) to EcoKI was 20:1. In each case the reaction mixture minus DNA was made up and incubated for 2 min at room temperature. The reaction was then initiated by adding DNA. Digestion was carried out for 8 min at 37°C before quenching at 68°C for 10 min. CC, closed circular plasmid; L, linear plasmid; OC, open circular plasmid.
KlcA136 does not bind to EcoKI Mtase
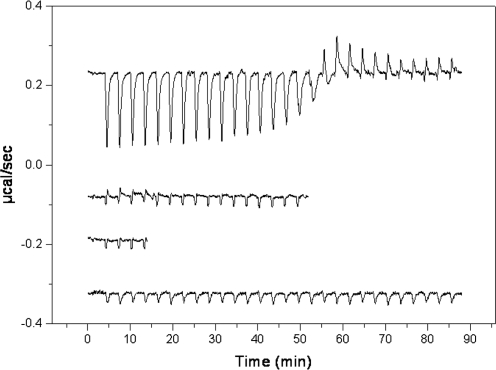
Biophysical characterization using ITC and fluorescence spectroscopy performed on two other anti-restriction proteins, ocr and ArdA, have shown a strong interaction of the Mtase part of EcoKI with these proteins (13,16). However, ITC experiments performed at 10 and 25°C showed no apparent binding of KlcA136 to Mtase, Figure 4. This result agrees with the in vivo modification assays where inhibition of phage DNA modification was not observed. Detecting the binding to the EcoKI nuclease was not feasible using ITC due to limited amounts of the nuclease. However, given the result of the nuclease inhibition assay, no interaction is expected with the surface of the nuclease.
Figure 4.
Raw ITC data showing heat changes for the interaction of KlcA136 with the EcoKI Mtase compared with data for the interaction of ArdA with the Mtase as previously published (13). The traces show from top to bottom: Mtase being titrated with ArdA at 25°C, buffer being titrated with Mtase at 25°C, Mtase being titrated with KlcA136 at 25°C and Mtase being titrated with KlcA136 at 10°C. It is clear that there is no interaction between the Mtase and KlcA136.
Do KlcA and ArdB interfere with assembly of EcoKI?
Since the KlcA and ArdB proteins appear unable to interact neither with the complete EcoKI RM enzyme nor with the core Mtase in vitro, we also examined whether they could interact with the surface of the HsdR restriction subunit of EcoKI that becomes buried on binding to the Mtase. This would interfere with the assembly of the complete RM enzyme as originally proposed by Belogurov and Delver (26). Addition of ArdB or KlcA prior to assembling the Mtase and HsdR into an intact EcoKI endonuclease failed to inhibit the endonuclease activity when DNA was subsequently added to the mixture, Figure 5. This strongly suggests that ArdB and KlcA do not interact with HsdR to interfere with assembly of EcoKI.
Figure 5.
The effect of adding ArdB or KlcA during the assembly of the EcoKI RM enzyme. Where relevant, Mtase and the R subunit were preincubated at 25°C in the presence of a × 20 excess of ArdB or KlcA protein for ∼5 min prior to mixing and addition of DNA. Reactions were performed at 37°C for 8 min. Lane 1, uncut pBRsk1 (3 nM); lane 2, pBRsk1 (3 nM) digested with EcoKI (30 nM); lane 3, pBRsk1 digested with reconstituted nuclease (equivalent to 30 nM EcoKI); lane 4, nuclease reconstituted in the presence of ArdBYAF (20-fold excess over nuclease); lane 5, 20-fold excess ArdBYFJ; lane 6, 20-fold excess KlcA136; lane 7, pBRsk1 in the presence of R subunit only; lane 8, pBRsk1 in the presence of Mtase only. M, 1 kbp size marker.
1H, 15N and 13C chemical shift assignments and structure calculations for KlcA136
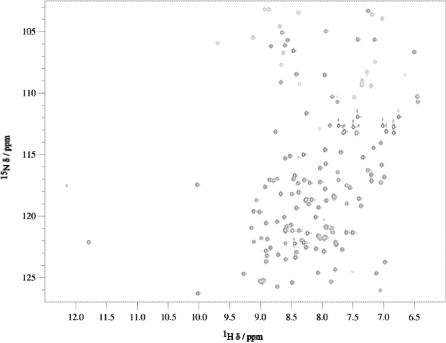
The recorded 1H-15N HSQC spectrum, Figure 6, was consistent with that of a folded protein exhibiting well dispersed signals and very little spectral overlap. Backbone and side-chain resonances of KlcA136 were assigned based on 2D and 3D triple-resonance experiments. Peaks in the 3D 13C and 15N edited NOESY spectra were picked and the NOE cross-peak assignments were obtained from the automated iterative assignment program CANDID that works in conjunction with 3D structure calculations in the program CYANA 2.1. Using NOE restraints, TALOS dihedral restraints and 1H-15N RDCs (Supplementary Figure S1), the lowest energy ensemble of 20 structures of KlcA136 were calculated (Supplementary Figure S2). The calculations converged well, yielding root mean square deviations of 0.63 and 0.65 for backbone and all heavy atoms in structured regions (residues 7–142). Only a subset of RDCs, as specified in the ‘Methods’ section for the calculation of the rotational diffusion tensor, was used in the structure calculations. The structures were validated using PROCHECK and WHATIF. In the Ramachandran plot, 98.5% of residues appeared in the most favourable and additionally allowed regions. WHATIF packing analysis yielded an average quality control z-score of −1.432 suggesting a good quality NMR structure.
Figure 6.
2D 1H-15N HSQC spectrum of KlcA136 at 800 MHz.
The structure of KlcA136 reveals a rigid globular protein with a novel fold
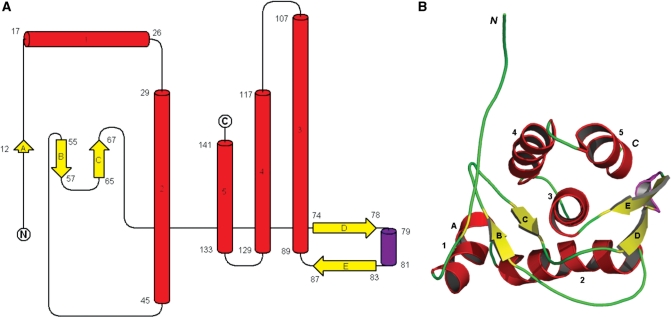
The KlcA136 protein structure is made up of a single domain (73) containing five α-helices (labelled 1–5), a single 310-helix and five β-strands (labelled A–E) and belongs to the mixed α/β class of proteins, Figure 7. The five β-strands form two separate beta-sheets, the first composed of anti-parallel β-strands A, B and C and the other made up of anti-parallel β-strands D and E. β-Strand A usually occurs as an isolated β-bridge from the ensemble consensus. The two β-sheets closely flank the C-terminal three α-helices (3–5) on either side. These three helices appear to run in an almost anti-parallel fashion to their helical axes in a classical knobs-in-holes arrangement of a three-helical bundle. Helix 3 is the longest of the helices located at the centre of the fold and contains several hydrophobic amino acids at its N-terminus that are deeply buried. A stereo view of the ensemble of the 20 lowest energy structures of KlcA136 is shown in Supplementary Figure S2.
Figure 7.
KlcA136 secondary and tertiary structure. Colour code: alpha-helices, red; beta-strands, yellow and 310 helix, purple. (A) Secondary structure elements identified by STRIDE for the ensemble consensus shown as a topology fold schematic (55–57). (B) A cartoon representation of the closest-to-mean 3D structure in the ensemble. Note: loops are rendered smooth for clarity.
Querying 3D-fold libraries and the PDB with the NMR-determined KlcA136 3D-structure demonstrated that its fold is novel, based upon its overall spatial arrangement and connectivity of secondary structure elements. For example, the strongest structural similarity found (z-score 5.1) using a DALI search of the PDB is to a discontinuous C-terminal portion of the 257-residue chain from G1/S-specific cyclin-D1 (PDB ID: 2W9Z), which is a purely α-helical protein. Although the z-score indicates structural similarity, the structurally aligned portions require a large 37-residue insertion between helices 2 and 3 (with respect to KlcA136) that encompass β-strands B–E, which are absent in the cyclin structure. Further confirmation of the novelty of fold was obtained using SSM. These results are consistent with the lack of sequence identity/significance scores for the B. pertussis KlcA sequence against PDB sequences using a BLAST search, or remote similarity to any existing folds, when undertaking fold recognition.
In the 1H-15N spectrum of KlcA136, two weak peaks were observed at 15N chemical shifts of 69 and 72 ppm. A F1 1H-coupled 1H-15N HSQC showed that these signals belong to NH2 moieties. The 15N chemical shifts are consistent with values reported for the guanidinium moiety of arginine from BioMagResBank (74). These NH2 groups would normally undergo chemical exchange with water and thus not be observed. The fact that they are present in the spectrum indicates reduced chemical exchange and suggests that these are involved in H-bonded salt bridges. Examination of the 3D structure of the protein, indicates that the Nη2 of R73 (one residue before strand D) and Oε2 of E86 (strand E) are separated by 2.97 Å, while this distance is 4.47 Å for R48 (loop) and E75 (strand D). The former pair meets the criterion (3.5 Å) for a H-bonded salt bridge interaction (75). Based on this analysis the 15N peaks with chemical shifts of 69 and 72 ppm were tentatively assigned to NH2 moieties of R48 and R73. Our relaxation data, described below, indicate that the D and E strands are the most flexible on the fast time scale, so a H-bonded salt bridge could stabilise these relatively short β-strands. The second interaction could potentially maintain the relative orientation between the helix 2 and strands D and E, which connect to the rest of the protein via loops.
The measured 15N longitudinal T1 and transverse T2 relaxation times and steady state 1H-15N NOEs were measured (Supplementary Figure S3). Due to spectral overlap, only 113 of the 137 non-proline residues of the protein could be used for the calculation of the relaxation parameters. The residues with higher amplitude of internal motions have simultaneously higher values of T1 and T2. In contrast, amide groups with the simultaneously lowest values of T1 and T2 values belong to the residues with the most restricted local mobility. Heteronuclear NOE values < 0.68 were used to identify residues of the protein that are dynamic on the ps–ns timescale. As the majority of heteronuclear NOEs are> 0.68, the backbone exhibits limited motions on this timescale (76). The only exceptions were residues 3–6 that exhibited negative NOE values indicating a flexible N-terminus.
The presence of internal motion and chemical exchange can lead to distortions of the T1/T2 ratio, and therefore the collection of residues used to determine the diffusion properties of the entire molecule excluded mobile residues or those exhibiting slow exchange broadening. A subset of 97 residues was used in the determination of the diffusion tensor. In this subset the average values of T1 and T2 were 737 and 86 ms, respectively. The relative moments of inertia values were found to be Iz: 1.00, Iy: 0.9858 and Ix: 0.8629. Iy and Ix are very close to 1.00, implying that the protein does not deviate significantly from a sphere, having slightly oblate shape. Given the nearly spherical shape, an isotropic diffusion tensor with a Diso = 1.8 × 107 s−1 was calculated and the average rotational correlation time (τc) was found to be 8.83 ns ± 0.01, a value expected for a protein of this size.
In order to extract microscopic parameters of motion from the relaxation data a model-free approach was used, which assumes the overall and internal motions of the molecule are independent. Based on the above analysis of the relaxation data, an isotropic model was selected. Relaxation data for most residues (73%) were adequately described by the generalized order parameter S2 suggesting a rigid molecule overall. The order parameter is a measure of the degree of spatial motion of the N–H bond where a value of zero means that the bond vector has unrestricted motion while one implies a completely rigid bond. An additional parameter (Rex) was required to fit 17 residues, with residues A14, A16, A91, E107, I108, L117 and Y121 showing significant Rex values (>2 Hz), suggesting that these N–H sites experience multiple environments interconverting on the micro- to millisecond time scale. Eleven residues (E4, M31, V38, W41, E47, Y56, F64, F99, Q103, F123 and A128) did not fit the model. The order parameter S2 as a function of the residue number is shown in Supplementary Figure S3. The distribution of S2 values visualized on the structure shows that the mobility on the nanosecond time scale is slightly restricted in secondary structure elements compared to loops (Supplementary Figure S4). The largest mobility indicated by the smallest S2 values was observed with residues at the N-terminus and with residues between the end of strand C and the start of helix 3.
The surface of KlcA136 reveals charged and hydrophobic patches but little sequence conservation for residues in these patches
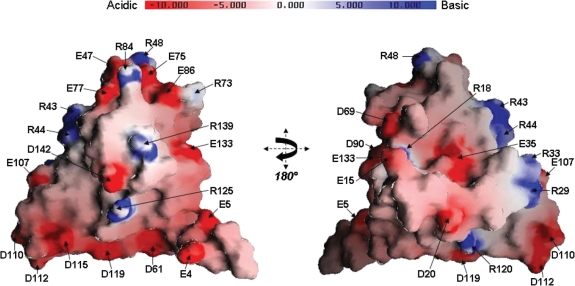
An electrostatic surface representation generated by GRASP shows that the surface of KlcA136 is predominantly negatively charged, consistent with its theoretical pI (4.76), Figure 8. The negatively charged side-chain residues on the KlcA136 surface reveals two distinct clusters (i) an ‘Aspartate cluster’ made up of five linearly arranged residues i.e. D61 (in the loop between β-strands B and C), D110, D112, D115, D119 (in the loop between α-helix 3 and 4 and on α-helix 4) and (ii) a ‘Glutamate cluster’ that contains amino acid residues E47 (in the loop between α-helix 2 and β-strand B), E75 and E77 (on β-strand D), E86 (on β-strand E) and E133 (on α-helix 5). In addition to the five residues present in the ‘Glutamate cluster’ are residues E15, D69 and D90, which are also located nearby in 3D space. A striking linear arrangement of positive charge involving arginine side-chains is also evident, predominantly on the opposite side of the KlcA136 protein surface with respect to its negatively charged clusters. This ‘Arginine cluster’ contains seven residues: R29, R33, R43, R44 on α-helix 2, R48 in the loop between α-helix 2 and β-strand B, R84 on β-strand E and R120 on α-helix 4. Even though three distinct regions of charge are apparent from the electrostatic surface representations, only a handful of the charged residues are conserved in the 43 non-redundant sequences identified (Supplementary Figure S5). These include R18, E35, D61, D90, R125, E133 and D142.
Figure 8.
Electrostatic surface representation of the KlcA136 protein. Two views rotated by 180° about the y-axis of a GRASP electrostatic surface representation of the KlcA136 protein. The molecule appears to expose many charged residues, labelled. Negative charge is coloured red and positive charge coloured blue, ranging from −10 kBT to +10 kBT (kB: Boltzmann constant; T: temperature in Kelvin).
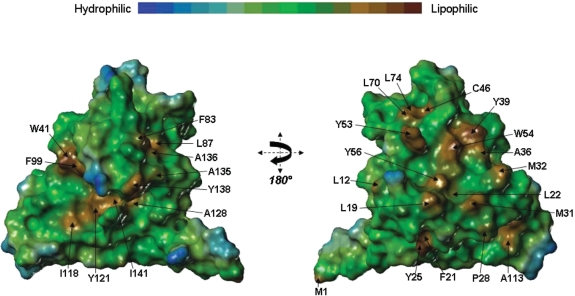
Additionally, the lipophilic surface rendition generated by MOLCAD reveals two regions of spatially contiguous hydrophobic side-chains on one face of the molecule, Figure 9 (left-hand panel) and another more prominent hydrophobic region on the opposite face of the protein, Figure 9 (right-hand panel). Among these, F21, L22, W54, F83, L87, A128 and I141 are largely conserved. Some of the aromatic side-chains are involved in aromatic–aromatic stacking interactions e.g., F64 with F21, Y25, F26; Y57 with Y65, F127; Y39 with W54 and W41 with F99.
Figure 9.
Lipophilic surface representation of the KlcA136 protein. Two views, rotated by 180° about the y-axis, of a MOLCAD-generated lipophilic surface rendition of the protein. Regions of high lipophilicity or hydrophobicity are coloured brown and regions of high hydrophilicity are coloured blue.
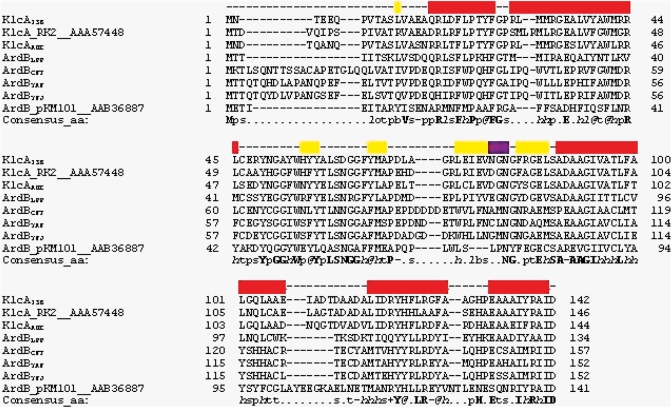
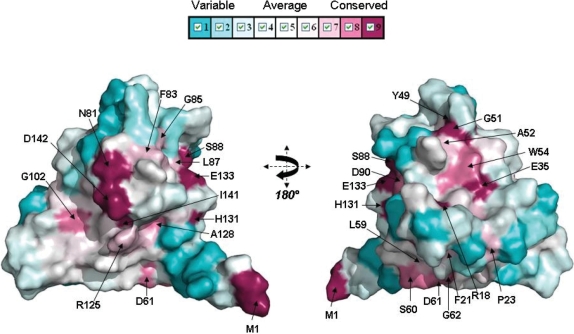
Comparisons between the 43 ArdB and KlcA protein sequences showed that they have highly conserved α-helices and β-strands, while loops vary in length and amino acid composition. Figure 10 shows a subset of the sequences comprised of the proteins listed in Table 1 and the archetypal KlcA from plasmid RK2 (20) and archetypal ArdB from pKM101 (11). Mapping sequence conservation (Supplementary Figure S5) onto the surface of KlcA136 shows only three well-conserved patches, comprising firstly H81 and D142, secondly L87, S88 and E133 and lastly E35, Y49, G51, A52 and W54, Figure 11. Only the first of these three patches projects out from the surface, the others are on concave regions of the surface.
Figure 10.
A PROMALS3D sequence alignment of KlcA and ArdB proteins. KlcA_RK2 and ArdB_pKM101 are the archetypal sequences for these two proteins (11,20). Yellow shading represents beta strands, red shading represents alpha helices and purple shading represents the 310 helix. The grey shading highlights the anti-restriction motif LLREYVNTL in ArdB from pKM101 (7,26). Consensus amino acid symbols are: conserved amino acids are in bold and uppercase letters; aliphatic (I, V, L): l; aromatic (Y, H, W, F): @; hydrophobic (W, F, Y, M, L, I, V, A, C, T, H): h; alcohol (S, T): o; polar residues (D, E, H, K, N, Q, R, S, T): p; tiny (A, G, C, S): t; small (A, G, C, S, V, N, D, T, P): s; bulky residues (E, F, I, K, L, M, Q, R, W, Y): b; positively charged (K, R, H): +; negatively charged (D, E): −; charged (D, E, K, R, H): c.
Figure 11.
Sequence conservation mapped onto KlcA136 protein surface. Conservation surface representation based upon the PROMALS3D multiple sequence alignment of ArdB and KlcA sequences (Supplementary Figure S5) where largely and completely conserved residues are shown in lavender/plum/violet colour mapped onto the surface and variable residue positions coloured cyan.
Discussion
In this study, we have solved the atomic resolution structure of KlcA136, an anti-restriction protein belonging to the ArdB family and showed its anti-restriction function against Type I RM systems. We have also demonstrated the same anti-restriction function for five other examples of the ArdB/KlcA family from a range of organisms.
A particularly striking result is the absence of any inhibition of a Type I RM enzyme in vitro. The EcoKI RM enzyme was able to bind and cleave unmodified DNA in the presence of either ArdB or KlcA proteins in contrast to the result in vivo in which the enzyme failed to operate against invading phage DNA in the presence of these anti-restriction proteins. Thus the ArdB/KlcA proteins must operate in a different manner to the DNA mimics. Several protein-based anti-restriction strategies aimed at negating the activity of Type I RM systems have been identified in addition to DNA mimicry (2), namely, hydrolysis of the essential S-adenosyl-l-methionine cofactor (77), proteolysis of the HsdR subunit required for DNA cleavage (30,32,71), enhancement of the Mtase activity via the action of the lambda ral gene to allow protection of unmethylated DNA targets (78) and interference with the subunit assembly process forming the complete RM enzyme (26). Our data and that of Belogurov et al. (11), who demonstrated that ardB did not stimulate DNA modification by the EcoKI Type I RM system using a ral-like mechanism, show that these mechanisms are not applicable to the ArdB/KlcA proteins. Thus the mechanism of anti-restriction by ArdB and KlcA remains unclear at present. However, the variable quaternary structure observed using size exclusion chromatography may suggest a role for large scale conformational changes in both tertiary and quaternary structure in ArdB and KlcA as being involved in anti-restriction.
The structure of KlcA136 is the first ArdB/KlcA structure reported and adopts a novel fold. Although three distinct regions of charge are apparent from the electrostatic surface representation, only a handful of charged residues are conserved among the 43 non-redundant related sequences of the ArdB/KlcA group. Other anti-restriction proteins, such as ocr from phage T7 (9) and ArdA from the conjugative transposon Tn916 (8), have been shown to comprise elongated structures that bind to DNA binding site of the Mtase core of the Type I RM enzyme, thereby rendering the enzyme incapable of recognizing its DNA target both in vivo and in vitro. The structure of KlcA136 on the other hand, appears to be a rigid globular molecule of novel fold and does not mimic double-stranded DNA.
ACCESSION NUMBER
2 kmg.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Biotechnology and Biological Sciences Research Council (BB/D001870/1 to D.T.F.D.); The Wellcome Trust (GR080463MA to D.T.F.D., G.W.B., Malcolm Walkinshaw and James Naismith, and G078780/Z/05/Z to D.U. and A.P.H.); The Engineering and Physical Sciences Research Council (studentship for D.S.M.); Wellcome Trust, BBSRC, Scottish Universities Life Science Alliance and The University of Edinburgh (to D.C.S. who is a part of Centre for Translational and Chemical Biology); RASOR grant from the BBSRC (BB/C511599/1 to Dr Patrick Langridge-Smith for access to the SIRCAMS FTICR-MS facility); Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Patrick Langridge-Smith for access to the SIRCAMS FTICR-MS facility supported by the RASOR grant from the BBSRC (BB/C511599/1) and Drs Logan Mackay and David Clarke for running the FT-ICR MS.
REFERENCES
- 1.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 2.Tock MR, Dryden DTF. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Wilkins BM. Plasmid promiscuity: meeting the challenge of DNA immigration control. Env. Microbiol. 2002;4:495–500. doi: 10.1046/j.1462-2920.2002.00332.x. [DOI] [PubMed] [Google Scholar]
- 4.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webb JL, King G, Ternent D, Titheradge AJ, Murray NE. Restriction by EcoKI is enhanced by co-operative interactions between target sequences and is dependent on DEAD box motifs. EMBO J. 1996;15:2003–2009. [PMC free article] [PubMed] [Google Scholar]
- 7.Zavilgelsky GB. Antirestriction. Mol. Biol. 2000;34:724–732. [Google Scholar]
- 8.McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, et al. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 2009;37:4887–4897. doi: 10.1093/nar/gkp478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DTF. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell. 2002;9:187–194. doi: 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 10.Stephanou AS, Roberts GA, Tock MR, Pritchard EH, Turkington R, Nutley M, Cooper A, Dryden DTF. A mutational analysis of DNA mimicry by ocr, the gene 0.3 antirestriction protein of bacteriophage T7. Biochem. Biophys. Res. Commun. 2009;378:129–132. doi: 10.1016/j.bbrc.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Belogurov AA, Delver EP, Rodzevich OV. Plasmid pKM101 encodes two nonhomologous antirestriction proteins (ArdA and ArdB) whose expression is controlled by homologous regulatory sequences. J. Bacteriol. 1993;175:4843–4850. doi: 10.1128/jb.175.15.4843-4850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nekrasov SV, Agafonova OV, Belogurova NG, Delver EP, Belogurov AA. Plasmid-encoded antirestriction protein ArdA can discriminate between type I methyltransferase and complete restriction-modification system. J. Mol. Biol. 2007;365:284–297. doi: 10.1016/j.jmb.2006.09.087. [DOI] [PubMed] [Google Scholar]
- 13.Serfiotis-Mitsa D, Roberts GA, Cooper LP, White JH, Nutley M, Cooper A, Blakely GW, Dryden DTF. The Orf18 gene product from conjugative transposon Tn916 is an ArdA antirestriction protein that inhibits type I DNA restriction-modification systems. J. Mol. Biol. 2008;383:970–981. doi: 10.1016/j.jmb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Thomas AT, Brammar WJ, Wilkins BM. Plasmid R16 ArdA protein preferentially targets restriction activity of the type I restriction-modification system EcoKI. J. Bacteriol. 2003;185:2022–2025. doi: 10.1128/JB.185.6.2022-2025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zavilgelsky GB, Kotova VY, Rastorguev SM. Comparative analysis of anti-restriction activities of ArdA (ColIb-P9) and Ocr (T7) proteins. Biochemistry. 2008;73:906–911. doi: 10.1134/s0006297908080087. [DOI] [PubMed] [Google Scholar]
- 16.Atanasiu C, Su TJ, Sturrock SS, Dryden DTF. Interaction of the ocr gene 0.3 protein of bacteriophage T7 with EcoKI restriction/modification enzyme. Nucleic Acids Res. 2002;30:3936–3944. doi: 10.1093/nar/gkf518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DTF. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 2009;37:762–770. doi: 10.1093/nar/gkn988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belogurov AA, Delver EP, Agafonova OV, Belogurova NG, Lee LY, Kado CI. Antirestriction protein Ard (Type C) encoded by IncW plasmid pSa has a high similarity to the “protein transport” domain of TraC1 primase of promiscuous plasmid RP4. J. Mol. Biol. 2000;296:969–977. doi: 10.1006/jmbi.1999.3493. [DOI] [PubMed] [Google Scholar]
- 19.Kamachi K, Sota M, Tamai Y, Nagata N, Konda T, Inoue T, Top EM, Arakawa Y. Plasmid pBP136 from Bordetella pertussis represents an ancestral form of IncP-1beta plasmids without accessory mobile elements. Microbiology. 2006;152:3477–3484. doi: 10.1099/mic.0.29056-0. [DOI] [PubMed] [Google Scholar]
- 20.Larsen MH, Figurski DH. Structure, expression, and regulation of the kilC operon of promiscuous IncP alpha plasmids. J. Bacteriol. 1994;176:5022–5032. doi: 10.1128/jb.176.16.5022-5032.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, et al. Escherichia coli K-12: a cooperatively developed annotation snapshot–2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasko DA, Phillips JA, Li X, Mobley HL. Identification of DNA sequences from a second pathogenicity island of uropathogenic Escherichia coli CFT073: probes specific for uropathogenic populations. J. Infectious Diseases. 2001;184:1041–1049. doi: 10.1086/323602. [DOI] [PubMed] [Google Scholar]
- 23.Welch RA, Burland V, Plunkett G, III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 25.Vedler E, Vahter M, Heinaru A. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 2004;186:7161–7174. doi: 10.1128/JB.186.21.7161-7174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belogurov AA, Delver EP. A motif conserved among the type I restriction-modification enzymes and antirestriction proteins: a possible basis for mechanism of action of plasmid-encoded antirestriction functions. Nucleic Acids Res. 1995;23:785–787. doi: 10.1093/nar/23.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamczyk M, Jagura-Burdzy G. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochimica Polonica. 2003;50:425–453. [PubMed] [Google Scholar]
- 28.Burlage RS, Bemis LA, Layton AC, Sayler GS, Larimer F. Comparative genetic organization of incompatibility group P degradative plasmids. J. Bacteriol. 1990;172:6818–6825. doi: 10.1128/jb.172.12.6818-6825.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harada KM, Aso Y, Hashimoto W, Mikami B, Murata K. Sequence and analysis of the 46.6-kb plasmid pA1 from Sphingomonas sp. A1 that corresponds to the typical IncP-1beta plasmid backbone without any accessory gene. Plasmid. 2006;56:11–23. doi: 10.1016/j.plasmid.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Blakely GW, Murray NE. Control of the endonuclease activity of type I restriction-modification systems is required to maintain chromosome integrity following homologous recombination. Mol. Microbiol. 2006;60:883–893. doi: 10.1111/j.1365-2958.2006.05144.x. [DOI] [PubMed] [Google Scholar]
- 31.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 32.Makovets S, Powell LM, Titheradge AJ, Blakely GW, Murray NE. Is modification sufficient to protect a bacterial chromosome from a resident restriction endonuclease? Mol. Microbiol. 2004;51:135–147. doi: 10.1046/j.1365-2958.2003.03801.x. [DOI] [PubMed] [Google Scholar]
- 33.Stephanou AS, Roberts GA, Cooper LP, Clarke DJ, Thomson AR, MacKay CL, Nutley M, Cooper A, Dryden DTF. Dissection of the DNA mimicry of the bacteriophage T7 Ocr protein using chemical modification. J. Mol. Biol. 2009;391:565–576. doi: 10.1016/j.jmb.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dryden DTF, Cooper LP, Thorpe PH, Byron O. The in vitro assembly of the EcoKI type I DNA restriction/modification enzyme and its in vivo implications. Biochemistry. 1997;36:1065–1076. doi: 10.1021/bi9619435. [DOI] [PubMed] [Google Scholar]
- 35.Uhrinova S, Uhrin D, Denton H, Smith M, Sawyer L, Barlow PN. Complete assignment of 1H, 13C and 15N chemical shifts for bovine beta-lactoglobulin: secondary structure and topology of the native state is retained in a partially unfolded form. J. Biomol. NMR. 1998;12:89–107. doi: 10.1023/a:1008268528695. [DOI] [PubMed] [Google Scholar]
- 36.Muhandiram DR, Farrow NA, Xu GY, Smallcombe SH, Kay LE. A gradient C-13 Noesy-Hsqc experiment for recording Noesy spectra of C-13-labeled proteins dissolved in H2O. J. Mag. Res. Series B. 1993;102:317–321. [Google Scholar]
- 37.Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-tailored water suppression for H-1-N-15 Hsqc experiments optimized to retain full sensitivity. J. Mag. Res. Series A. 1993;102:241–245. [Google Scholar]
- 38.Yamazaki T, Forman-Kay JD, Kay LE. Two-dimensional NMR experiments for correlating 13Cβ and 1Hδ/ε chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J. Am. Chem. Soc. 1993;115:11054–11055. [Google Scholar]
- 39.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 40.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 41.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 42.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 45.Vriend G. What if - a molecular modeling and drug design program. J. Mol. Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 46.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55, 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 47.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 48.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. J. Magn. Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Grzesiek S, Bax A. The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 50.Yip GN, Zuiderweg ER. Improvement of duty-cycle heating compensation in NMR spin relaxation experiments. J. Magn. Reson. 2005;176:171–178. doi: 10.1016/j.jmr.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 52.Tjandra N, Feller SE, Pastor RW, Bax A. Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J. Am. Chem. Soc. 1995;117:12562–12566. [Google Scholar]
- 53.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 55.Hutchinson EG, Thornton JM. HERA–a program to draw schematic diagrams of protein secondary structures. Proteins. 1990;8:203–212. doi: 10.1002/prot.340080303. [DOI] [PubMed] [Google Scholar]
- 56.Hutchinson EG, Thornton JM. PROMOTIF–a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bond CS. TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics. 2003;19:311–312. doi: 10.1093/bioinformatics/19.2.311. [DOI] [PubMed] [Google Scholar]
- 58.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 60.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 61.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 62.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 64.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 66.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 67.Heiden W, Moeckel G, Brickmann J. A new approach to analysis and display of local lipophilicity hydrophilicity mapped on molecular-surfaces. J. Comput. Aided Mol. Des. 1993;7:503–514. doi: 10.1007/BF00124359. [DOI] [PubMed] [Google Scholar]
- 68.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comp. Chem. 1998;19:319–333. [Google Scholar]
- 69.Saltman LH, Kim KS, Figurski DH. The kilA operon of promiscuous plasmid RK2: the use of a transducing phage (lambda pklaA-1) to determine the effects of the lethal klaA gene on Escherichia coli cells. Mol. Microbiol. 1991;5:2673–2683. doi: 10.1111/j.1365-2958.1991.tb01976.x. [DOI] [PubMed] [Google Scholar]
- 70.Dunn JJ, Elzinga M, Mark KK, Studier FW. Amino acid sequence of the gene 0.3 protein of bacteriophage T7 and nucleotide sequence of its mRNA. J. Biol. Chem. 1981;256:2579–2585. [PubMed] [Google Scholar]
- 71.Makovets S, Doronina VA, Murray NE. Regulation of endonuclease activity by proteolysis prevents breakage of unmodified bacterial chromosomes by type I restriction enzymes. Proc. Natl Acad. Sci. USA. 1999;96:9757–9762. doi: 10.1073/pnas.96.17.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bandyopadhyay PK, Studier FW, Hamilton DL, Yuan R. Inhibition of the type I restriction-modification enzymes EcoB and EcoK by the gene 0.3 protein of bacteriophage T7. J. Mol. Biol. 1985;182:567–578. doi: 10.1016/0022-2836(85)90242-6. [DOI] [PubMed] [Google Scholar]
- 73.Alexandrov N, Shindyalov I. PDP: protein domain parser. Bioinformatics. 2003;19:429–430. doi: 10.1093/bioinformatics/btg006. [DOI] [PubMed] [Google Scholar]
- 74.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar S, Nussinov R. Salt bridge stability in monomeric proteins. J. Mol. Biol. 1999;293:1241–1255. doi: 10.1006/jmbi.1999.3218. [DOI] [PubMed] [Google Scholar]
- 76.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 77.Studier FW, Movva NR. SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J. Virol. 1976;19:136–145. doi: 10.1128/jvi.19.1.136-145.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loenen WA, Murray NE. Modification enhancement by the restriction alleviation protein (Ral) of bacteriophage lambda. J. Mol. Biol. 1986;190:11–22. doi: 10.1016/0022-2836(86)90071-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.