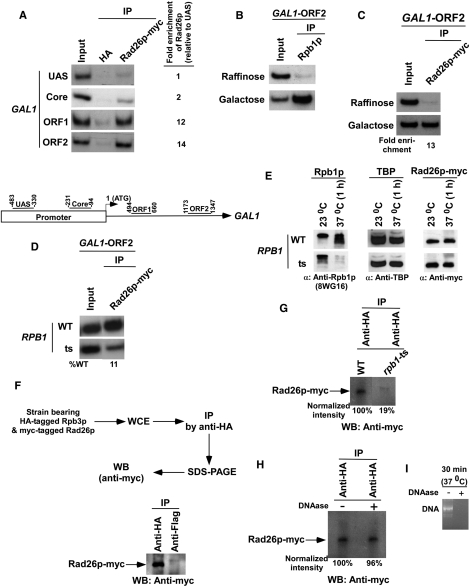
Figure 1.
Rad26p is associated with the coding sequence but not promoter of the GAL1 gene in a transcription-dependent manner. (A) Top panel: analysis of the recruitment of Rad26p at the UAS, core promoter, ORF of the GAL1 gene. The yeast strain expressing myc-tagged Rad26p was grown in YPG up to an OD600 of 1.0 prior to cross-linking. The ChIP assay was performed as described in the Materials and Methods section. Immunoprecipitation was performed using a mouse monoclonal antibody against the c-myc epitope-tag (9E10; Santa Cruz Biotechnology, Inc.). An anti-HA (Santa Cruz Biotechnology, Inc.) was used as a non-specific antibody. Both the input and immunoprecipitated DNA samples were analyzed by PCR. The dilution factors for input and immunoprecipitated DNA samples are mentioned in the modified ChIP protocol (see ‘Materials and Methods’ section). The immunoprecipitated DNA was quantitated as the ratio of immunoprecipitate over the input in the autoradiogram. The fold enrichment of Rad26p at the GAL1 coding sequence with respect to UAS is presented. IP, immunoprecipitate. Bottom panel: the PCR primer pairs located at the UAS, core promoter, and towards the 5′-(ORF1) and 3′-(ORF2) ends of the ORF of the GAL1 gene. (B) RNA polymerase II is associated with the coding sequence of the active GAL1 gene. The yeast strain was grown in raffinose (YPR)- or galactose (YPG)-containing growth medium up to an OD600 of 1.0 prior to crosslinking. Immunoprecipitations were performed using a mouse monoclonal antibody 8WG16 (Covance) against the carboxy terminal domain of the largest subunit (Rpb1p) of RNA polymerase II. (C) Rad26p is associated with the ORF of the active GAL1 gene. The fold enrichment of Rad26p at the GAL1 coding sequence in galactose-containing growth medium in comparison to raffinose-containing growth medium is presented. (D) Rpb1p is essential for recruitment of Rad26p to the GAL1 coding sequence. (E) Analysis of the global levels of Rad26p in the rpb1-ts and wild-type strains at the non-permissive temperature. The yeast strain expressing myc epitope-tagged Rad26p were grown in YPD medium at permissive (23°C) and non-permissive (37°C) temperatures as discussed in the ‘Materials and Methods’ section. The WCE was run on SDS–polyacrylamide gel, and then analyzed by western blot assay. (F) Rad26p interacts with RNA polymerase II as revealed by the co-immunoprecipitation assay. (G) Analysis of Rad26p association with RNA polymerase II in the rpb1-ts mutant using a co-immunoprecipitation assay. (H) The co-immunoprecipitation assay as in panel G in the presence of DNAase. (I) Analysis of DNA digestion as a control for the experiments presented in (H). PCR-amplified DNA was treated with DNAase for 30 min at 37°C, and then was analyzed by agarose gel electrophoresis.