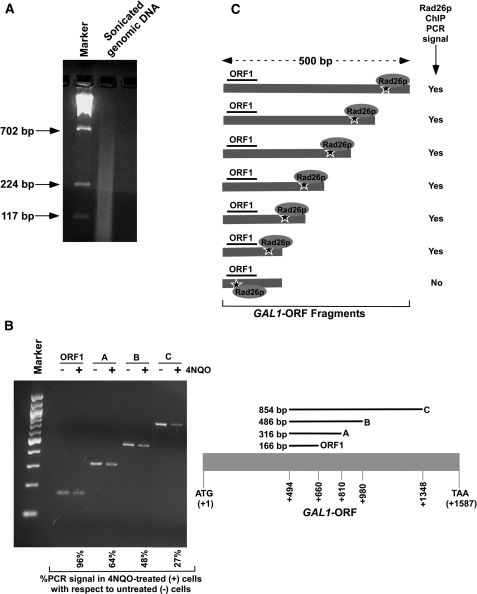
Figure 4.
Analysis of DNA lesion in the GAL1 coding sequence. (A) Analysis of the size of DNA fragments in the ChIP assay. Yeast cells were grown in YPR prior to cross-linking as in Figure 3B. The genomic DNA was isolated following sonication (7 times, 10 s each) of WCE and was analyzed by agarose gel electrophoresis. (B) Analysis of DNA damage at GAL1 with varying lengths of coding sequence. Yeast cells were grown, cross-linked and sonicated as in (A). Cells were treated with 4NQO at a final concentration of 4 µg/ml as described in Figure 3A. DNA was isolated and analyzed by PCR using specific primer pairs. PCR products with different sizes were analyzed by 2% agarose gel electrophoresis. (C) The schematic diagram for the analysis of Rad26p recruitment at the site of DNA lesion in the ChIP assay. The ‘star’ represents the site of DNA lesion.