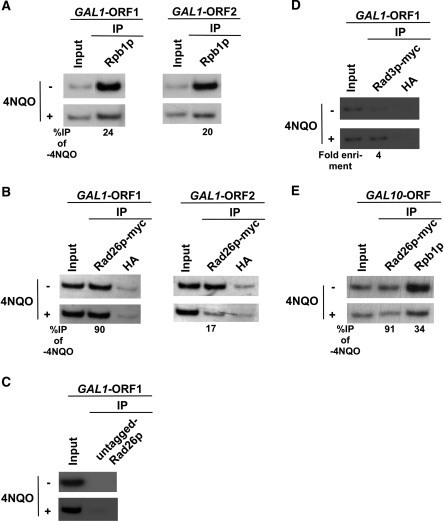
Figure 5.
RNA polymerase II promotes the recruitment of Rad26p to the DNA lesions at the coding sequences of the active GAL1 and GAL10 genes. (A) Analysis of the association of RNA polymerase II with the GAL1 coding sequence in the presence and absence of 4NQO-induced DNA damage. The yeast strain was grown in YPR up to an OD600 of 0.8 at 30°C, and then transferred to YPG for 90 min to induce GAL1 prior to 4NQO treatment for 20 min. As a control, yeast cells were also grown under similar growth conditions without 4NQO treatment. These cells were used for the ChIP assay to analyze the level of Rpb1p at the GAL1 ORF in 4NQO-treated (+) and untreated (−) cells. The percentage of DNA immunoprecipitated in the 4NQO-treated cells relative to that of the untreated cells is represented as ‘%IP of -4NQO’. (B) Analysis of Rad26p recruitment towards the 5′- and 3′-ends of the GAL1 ORF in the presence and absence of 4NQO-induced DNA damage. The yeast strain carrying myc-tagged Rad26p was grown and crosslinked as in (A). Immunoprecipitation was performed as in Figure 1A. (C) The ChIP experiments at the GAL1 coding sequence using the strain that bears untagged Rad26p. An anti-myc antibody was used in the ChIP assay. (D) Analysis of Rad3p recruitment at the GAL1 coding sequence following 4 µg/ml 4NQO treatment. Immunoprecipitation was performed using the modified ChIP protocol as described in the ‘Materials and Methods’ section. (E) Analysis of the association of Rad26p and RNA polymerase II with the GAL10 ORF.