Abstract
The spliceosomal protein Prp1 (Prp6/U5-102 K) is necessary for the integrity of pre-catalytic spliceosomal complexes. We have identified a novel regulatory function for Prp1. Expression of mutations in the N-terminus of Prp1 leads to the accumulation of pre-catalytic spliceosomal complexes containing the five snRNAs U1, U2, U5 and U4/U6 and pre-mRNAs. The mutations in the N-terminus, which prevent splicing to occur, include in vitro and in vivo identified phosphorylation sites of Prp4 kinase. These sites are highly conserved in the human ortholog U5-102 K. The results presented here demonstrate that structural integrity of the N-terminus is required to mediate a splicing event, but is not necessary for the assembly of spliceosomes.
INTRODUCTION
The chemical reactions leading to the removal of introns from pre-mRNAs and the basic components, including the five small nuclear ribonucleoproteinparticles (U1, U2, U5 and U4/U6 snRNPs) of a spliceosome, are conserved throughout evolution (1). Two transesterification reactions are catalyzed by this very dynamic snRNP–protein machinery. A series of conformational changes is induced to drive a pre-catalytic spliceosome into catalysis. These conformational changes are important for splicing regulation and quality control (2,3).
A spliceosome assembles in vitro on a pre-mRNA in a step-wise manner. The snRNPs U1 and U2 recognize the 5′- and 3′-splice sites, respectively, and then a tri-snRNP consisting of U4/U6.U5 is recruited to build up the pre-catalytic spliceosomal complex containing the five U snRNPs (2). It is not known how a splicing competent spliceosome bound to its substrate initiates the switch towards the multiple rearrangements to form a catalytic complex. However, it has been shown that the pre-catalytic spliceosome releases U1 and U4 to rearrange into the catalytic subcomplex containing the snRNAs U2.U6.U5 (2,4). How assembly and activation of the spliceosomal machinery occurs in vivo is still a matter of intense debate (5). It has been suggested that assembly of spliceosomes in vivo as well might be a stepwise process (6–8). However, the isolation and characterization of a spliceosomal complex from the budding yeast Saccharomyces cerevisiae containing the five U snRNAs and most of the known snRNP proteins led to the suggestion that a penta-snRNP (holospliceosome) may assemble independently of pre-mRNA (9). The results of other experimental approaches to determine the sequential order of events in forming a splicing competent spliceosome in vivo suggest that the U1 snRNP recognizes pre-mRNA first. However, it cannot be decided whether in the next step U2 and then a pre-formed tri-snRNP U4/U6.U5 or a tetra-snRNP U2.U4/U6.U5 is recruited to the U1 snRNP-pre-mRNA complex (7–11).
Almost nothing is known how and when an assembled spliceosome is switched on for catalysis in vivo. The proteins involved in this switch and, particularly, their specific function remains unknown. We have shown that Prp4 kinase and its physiological substrate Prp1 play a regulatory role in pre-mRNA splicing in fission yeast Schizosaccharomyces pombe. This interaction appears to be conserved throughout eukaryotic organisms, with the exception of S. cerevisiae (12–14). Mammalian PRP4K kinase was found associated with spliceosomes and spliceosomal proteins, including U5-102 K the ortholog of Prp1 (15,16). The ortholog of Prp1 in budding yeast S. cerevisiae is called Prp6. In S. pombe prp1+ cannot be replaced by the S. cerevisiae PRP6 gene and the budding yeast genome does not encode an ortholog of Prp4 kinase (17,18).
Prp1/Prp6/U5-102 K is a protein consisting of multiple direct repeats formerly called TPRs (Tetra-trico Peptide Repeats), which are now listed as HATs (half a TPR) in databases (Q12381, PRP1_SCHPO; P19735, PRP6_YEAST; O94906, PRP6_HUMAN). In Prp1 11 HAT repeats are preceded by a 27-kDa domain at the N-terminus of the protein which does not show similarities with known motifs. Interestingly, whereas the C-terminus displaying the HAT repeats is highly conserved among organisms, the N-terminus is less conserved.
In S. cerevisiae Prp6 is necessary for the formation of a stable tri-snRNP U4/U6.U5. It was suggested that both yeast and human Prp6 (U5-102 K) are required for tri-snRNP integrity, whereas Prp6 and Prp31 are connecting the U5 snRNP with the U4/U6 snRNP particle (19–24).
Prp1 is essential for growth and expressed by a single gene on chromosome II. Different temperature sensitive (ts) alleles of prp1 accumulate pre-mRNA and the cell population arrests specifically either in G1 or in G2 phase of the cell cycle. This phenotype is caused by point mutations in the C-terminal part (HAT motifs) of Prp1 (12,14,17).
Attempts to study the stepwise assembly of spliceosomal particles with S. pombe whole-cell extracts failed, due to the lack of an in vitro splicing system. These investigations revealed, however, that whole-cell extract of S. pombe contains minute amounts of tri-snRNP U4/U6.U5 (25).
Three in vivo derived large spliceosomal complexes from S. pombe have been isolated and analyzed. First, we have isolated and analyzed the U1 particle. Unexpectedly, the U1 snRNP exists as a multimeric 45S particle containing exclusively snRNA U1 associated with the seven Sm core proteins and nine U1 specific proteins (26). Second, based on the analysis of a 35S complex containing the snRNAs U2, U5 and U6, it was shown that it consists of a mixture of activated- and post-catalytic spliceosomal complexes. These spliceosomal subcomplexes did not contain the bona fide splicing factors Prp1/Prp6/U5-102 K and Prp31 (27–30). Third, a complex associated with Prp1/Prp6/U5-102 K and Prp31 has been isolated and proteomic analysis of the complex revealed that it contains proteins expected in a spliceosomal sub-complex containing the four snRNAs U2, U5 and base paired U4/U6 (31). Prp1 and Prp31 are found associated with large spliceosomal complexes co-sedimenting in a glycerol gradient in the range of 30–60S. The spliceosomal complexes sedimenting in this range appear to be a mixture of pre-catalytic spliceosomal particles containing the four snRNAs U2, U5 and U4/U6 as well as complexes containing the five snRNAs, including U1. Collectively, these results indicate that Prp1 and Prp31 are proteins associated with pre-catalytic spliceosomal particles, but not with post-catalytic complexes (14,28,30,31).
Based on the observations described earlier, we hypothesized that Prp1 might play a regulatory role in splicing and is involved in the integrity and activation of pre-catalytic spliceosomal complexes.
Here, we report that expression of Prp1 carrying mutations in the N-terminus leads to the accumulation of pre-catalytic spliceosomal complexes containing pre-mRNAs. Spliceosomal complexes associated with mutant Prp1 appear to stall in a pre-catalytic stage containing all five snRNPs U1, U2, U5 and U4/U6. The results presented here indicate that mutations in the N-terminus of Prp1 prevent activation, but not assembly of pre-catalytic spliceosomes.
MATERIALS AND METHODS
Schizosaccharomyce pombe strains, growth media and in vivo tagging
Standard genetic and molecular techniques were used as described earlier (14). Strains were crossed and grown in media as described (32). All genotypes of the strains used in this study are listed in Table 1 .
Table 1.
Schizosaccharomyces pombe strains used in this study
| Strain | Genotype |
|---|---|
| L972 | h−S |
| 425 | h−S leu1-32 int::pJK148nmt1-8 prp1 ura4-294 prp1::his7 his7-366 prp31 int::HA-prp31 |
| 470 | h−S leu1-32 int::pJK148nmt1-8 prp1 ura4-294 int::pUR19-MycPrp1 prp1::his7 his7-366 prp31 int::HA-prp31 |
| 714 | h−S leu1-32 int::pJK148nmt1-8 prp1 ura4-294 int::pUR19-MycPrp1Δ227-249 prp1::his7 prp31 int::prp31-CTAP-KanMx6 his7-366 |
| 497 | h−S leu1-32 int::pJK148nmt1-8 prp1 ura4-294 int::pUR19-MycPrp1Δ227-249 prp1::his7 prp31 int::HA-prp31 his7-366 |
For construction of strain 425, prp1 was fused to the thiamine-repressible nmt1-8 promotor by cloning the prp1 cDNA into the vector pRIP81 (33,34). The nmt1-8 promotor, prp1 cDNA and nmt1-8 termination sequence were subsequently cloned as a cassette into the plasmid pJK148 (35). This construct (pJK148nmt1-8 prp1) was linearized and integrated into the leu1 locus by homologous recombination. Prototrophic transformants were checked for integration by polymerase chain reaction (PCR). In transformants, harboring the nmt1-8 controlled prp1 cDNA, the prp1 locus was disrupted by integration of the his7 gene, using a construct containing flanking sequences of 300 bp both upstream and downstream of the prp1 ORF.
To construct a Myc-tagged version of Prp1, we joined in a pUR19 vector (36) three PCR fragments comprising the promoter region of prp1, a cassette comprising a Myc-epitope and a fragment containing the ORF and 3′ UTR of prp1. For integration via homologous recombination into the ura4 locus, the plasmid was linearized in the ura4 gene and transformed into strain 425 auxotroph for uracil, containing the ura4-294 allele. Strains expressing epitope tagged versions of Prp31 were constructed by introducing a CTAP-KanMX6 or 3× HA-Prp31 module by homologous recombination at the 3′- and 5′-end of the endogenous locus, respectively (37,38). Expression of epitope-tagged proteins was confirmed by immunoblotting using the appropriate antibodies (39).
Tap tag purification of spliceosomal complexes
Prp31-CTAP containing spliceosomal complexes were purified from strain 714 as described by Gould et al. (40) with several modifications of the protocol (26). Cells were grown in 5 l Edinburgh Minimal Medium (EMM) at 30°C in absence or presence of 200 µM thiamine and were harvested after 16–18 h. The cell pellet (∼30 g fresh wet weight) was frozen in liquid nitrogen and stored at –80°C until use. As a negative control, strain 497, expressing Prp31-HA and Myc-Prp1 was cultivated under the same growth conditions. For protein extract preparation, cells were disrupted in a bead beater (BioSpec Products) in 20 ml of NP-40 buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 1% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 4 µg/ml leupeptin, 0.1 mM Na3VO4), containing protease inhibitors (1×Complete Mini EDTA-free, Roche Diagnostics, 1.3 mM, benzamidin, 1 mM PMSF) and RNase inhibitor (20 mM vanadyl ribonucleoside complex), prepared after Berger and Birkenmeier (41). After cell disruption, glass beads were washed with an additional 20 ml of NP–40 buffer. The lysate was cleared by centrifugation at 16 000g for 15 min at 4°C and immediately used for purification. The extract was split into four aliquots of 10 ml and placed into 0.8 × 4 cm chromatography columns (Bio-Rad, Poly-Prep), each supplied with 300 µl bed volume of IgG-Sepharose beads (GE Healthcare, IgG-Sepharose 6, Fast Flow) equilibrated in NP-40 buffer.
The beads were incubated on a rotating wheel at 4°C for 2 h and subsequently washed three times with 10 ml of IPP150 buffer (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.1% NP-40) and three times with 1 ml of TEV cleavage buffer (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1.0 mM DTT). After the last washing step, the beads were resuspended in 300 ml of TEV cleavage buffer. Then 40 U recombinant TEV protease (MoBiTec GmbH, Goettingen) were added and the beads were incubated on a rotating wheel at 18°C for 3 h.
After incubation with TEV protease, about 1300 µl of the eluate were collected. Three-hundred microliters of the eluate were mixed with 100 µl of 100% TCA and the precipitated proteins were analyzed by western blotting and Coomassie staining. To the remaining eluate (about 1000 µl) 3 ml calmodulin binding buffer (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, 1 mM DTT) were added and incubated with 300 µl calmodulin affinity resin (Stratagene) on a rotating wheel for 1 h at 4°C. The beads were washed with 3 ml calmodulin binding buffer containing 0.1% NP-40 and 2 ml calmodulin binding buffer containing 0.02% NP-40. The resin bound complexes were eluted either under native conditions by resuspending the resin in 1000 µl calmodulin elution buffer (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.02% NP-40, 20 mM EGTA, 1 mM DTT) or denaturing conditions using calmodulin elution buffer containing 0.1% sodium dodecyl sulfate (SDS).
About 300 µl of the eluate were used for TCA precipitation. Precipitated proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and subsequent western analysis and/or Coomassie staining. From the remaining eluate (700 µl), RNA was isolated by phenol/chloroform extraction and ethanol/sodium acetate precipitation.
Protein extracts
Protein extracts were prepared after a protocol published by Moreno et al. (39). In brief, cells were harvested and washed in STOP buffer (150 mM NaCl, 50 mM NaF 10 mM EGTA, 1 mM NaN3, pH 8.0). The cell pellet was resuspended in HB buffer (25 mM MOPS pH 7.2, 15 mM EGTA, 15 mM MgCl2, 150 mM NaCl, 60 mM β-glycerophosphate, 1 mM DTT, 0.1 mM Na3VO4) containing protease inhibitors (1 × Complete Mini EDTA-free, Roche Diagnostics, 4 µg/ml leupeptin, 1.3 mM, benzamidin, 1 mM PMSF) and disrupted with 0.3 mm glass beads in a bead beater (FP120 FastPrep cell disrupter; Savant Instruments, Inc.). After centrifugation (13 800g at 4°C) for 15 min, the supernatant was frozen in liquid nitrogen and stored at –80°C until use.
Glycerol gradient centrifugation
About 3.5–7 mg of total protein extract was loaded onto an 11 ml 10–30% glycerol gradient in HB buffer, and spun at 24 500 r.p.m. for 15 h at 4°C using a SW-41 rotor (Beckman). Nineteen 500-ml fractions were collected manually from top to bottom. Ten microliters of each fraction were subjected to solution hybridization. 30S and 50S ribosomal subunits from Escherichia coli were used to calibrate the gradients.
Solution hybridization of snRNA
Spliceosomal snRNAs were detected by a solution hybridization assay as described by Li and Brow (42). The samples were incubated in 50 ml SHS buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA pH 8.0, 0.1% SDS, 1 mg/ml proteinase K) for 30 min at 37°C, extracted with phenol/chloroform/isoamyl alcohol (50: 48: 2) and ethanol precipitated. The RNA was hybridized with 0.1 pmol [32P]-labeled oligonucleotides complementary (c) to U1, U2, U4, U5 and U6 for 30 min at 37°C. After addition of 6× loading buffer (50% glycerol, 10 mM EDTA pH 8.0, 0.025% bromophenol blue, 0.025 % xylene cyanol), the RNA was separated on a native polyacrylamide gel (9%; 30 : 1 acrylamide/bisacrylamide, 0.5× TBE) with constant power (5 W) and temperature (48°C). Oligonucleotides used: cU1 (1–26) 5′-GCTGCAGAAACTCATGCCA-GGTAAGT-3′; cU2 (35–55) 5′-GAACAGATACTACACTTGATC-3′; cU4 (71–92) 5′-GTTGGAGCGGTCAGGGTAATAG-3′; cU5 (77–107) 5′-GATTACAAAAAC-TATACAGTCAAATTAGCAC-3′; cU6 (18–40) 5′-CTCTGTATCGTTTCAATTTGACC-3′.
Reverse transcriptase-PCR
Prior to Reverse transcriptase (RT)-PCR, the RNA extracted from TAP eluates was treated with RQ1 RNase-free DNase (Promega) in order to eliminate possible DNA contaminants. Each microgram of RNA was treated with 0.1 U RQ1 DNase in 1× reaction buffer [40 mM Tris–HCl (pH 8.0), 10 mM MgSO4 and 10 mM CaCl2]. After incubation for 30 min at 37°C, the RQ1 DNase was inactivated by the addition of 2 mM EGTA, pH 8.0 and heating to 65°C for 10 min.
The RNA was reverse transcribed, and the cDNA amplified with MasterAmp™ Tth polymerase (Epicentre) using the ‘single-tube’ protocol according to the manufacturer’s instructions. In brief, the RNA was first incubated in presence of 1× reaction buffer (20 mM NH4SO4, 50 mM Tris–HCl pH 9.0), 3 mM MgCl2, 400 µM dNTP (each), 1× MasterAmp™ PCR enhancer, 0.5 mM MnSO4, 0.5 µM of forward and reverse primer and 1.25 U MasterAmp™ Tth polymerase for 20 min at 65°C. Then, the cDNA was amplified with 18–25 cycles of 94°C for 30 s, 54°C for 30 s and 72°C for 25 s. The PCR products were separated on a 3% agarose gel using 1× TBE as a running buffer and stained with ethidium bromide.
For amplification of the U snRNAs, the following primers were used: U1F 5′-CGGGATCCACTTACCTGGCATGAGTTTC-3′, U1R 5′-ACGCGTCGACTGCCCCAAATGAGGGACG-3′, U2F 5′-ATATGGATCCATTCTCTCTTTGCCTTTTG-3′, U2R 5′-ATATGTCGACATTCGGCGTCGCTTGCCA-3′, U4F 5′-ATATGGATCCATCTTTGTGCACGGGTATT-3′, U4R 5′-ATATGTCGACAGTTGGTTTCCAAATATTCC-3′, U5F 5′-ATATGGATCCATAATCCGTCAAAGCACTTT-3′, U5R 5′-ATATGTCGACTTTCAAGAAAAAGATTACAAAAA-3′, U6F 5′-ATATGGATCCGATCTTCGGATCACTTTGG-3′, U6R 5′-ATATGTCGACAAAATGGGTTT-TCTCTCAAT-3′.
For amplification of transcripts revealing spliced and unspliced versions of the ribosomal protein genes rpl29 and rps27, the following primers were used: rpl29F 5′-ATGGCCAAGTCGAAGAATCATACTA-3′, rpl29R 5′-TTGGTTGCGGCGGAACTTA-3′, rps27F 5′-TACCCCGAGCAATTCACTAACA-3′, rps27R 5′-ACAACGGTTTGAGCATGAGAGA-3′.
Immunoprecipitation
For immunoprecipitation assays, 5 µg of monoclonal antibodies (HA.11, Hiss Diagnostics) bound to protein A sepharose (Sigma-Aldrich) was incubated with 500 µl of each gradient fraction at 4°C for 2–16 h. The immunoprecipitates were washed two times with 1 ml buffer containing 150 mM NaCl, 10 mM Tris–HCl (pH 7.5) and 0.05% NP-40 at 4°C.
For immunoblotting, 50% of the immunoprecipitates were separated on an SDS-PAGE and transferred onto Protran nitrocellulose membrane (Whatman) by semi-dry blotting, according to the manufacturer’s instructions. After blocking over night with 10% non-fat milk powder in 1× TBS, the membranes were incubated with anti-HA antibodies. After washing with 1× TBS, the membranes were incubated with the HRP-conjugated anti-mouse antibodies (Amersham Biosciences). Protein bands were visualized by chemiluminescence (Pierce SuperSignal West Pico substrate) according to the manufacturer’s instructions. To detect snRNA, 50% of the immunoprecipitates were incubated in 50 µl SHS buffer as described earlier.
Expression and purification of recombinant Prp4 and Prp1
Prp4 was expressed in E. coli DH5α as a GST fusion protein by cloning the prp4 cDNA into the plasmid pGEX-6P-1 (GE Healthcare). For production of recombinant Prp4, 10 ml of an over night pre-culture were diluted into 500 ml of LB medium, incubated for 2 h at 37°C and induced with 1 mM IPTG for 3 h at 37°C. Cells were harvested by centrifugation and washed with 1× PBS (137 mM NaCl; 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.4). The cell pellet was frozen in liquid nitrogen and stored at –80°C until use. Cells were disrupted by sonication (five pulses of 10 s, 100 W) in 1.5 ml lysis buffer (1 × TBS, containing 1 mg/ml lysozyme, 30 µg/ml RNase A, 30 µg/ml DNase I, 1 mM PMSF and 1× Complete Mini, EDTA-free protease inhibitor). After clarification of the lysate by centrifugation with 16 000g for 20 min, 600 µl glutathione sepharose (4B, GE Healthcare) were added and incubated for 45 min at 4°C with agitation. The sedimented Sepharose beads were washed with 30 bed volumes of 1× PBS containing 1% (v/v) Triton X-100, 1 mM DTT; and with 10 bed volumes GST cleavage-buffer (50 mM Tris–HCl pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT). To elute recombinant Prp4, the beads were treated with 100 U PreScission protease (GE Healthcare) in 600 µl GST cleavage-Puffer for 4 h at 4°C on a rotating wheel. The eluate was concentrated by ultra filtration (Vivaspin 0.5 ml, 10 000 MWCO, Sartorius) and stored in kinase buffer [20 mM HEPES-KOH pH 7.4, 3 mM MgCl2, 5% (v/v) glycerol, 0.5 mM DTT] at –80°C until use.
Recombinant His6-Prp1 was expressed in E. coli M15[pREP4] by cloning the prp1 cDNA into pQE31 (Qiagene). For induction, an over night pre-culture of 50 ml was diluted into 500 ml LB medium, incubated at 37°C until an OD600 of 0.5 was reached, chilled to 17°C and incubated for 16 h in presence of 1 mM IPTG. After harvesting, cells were washed in sodium phosphate buffer (50 mM Na2HPO4, pH 7.4, 300 mM NaCl) and lysed by sonication in lysis buffer (50 mM Na2HPO4, 300 mM NaCl, 10 mM imidazol, 1 mg/ml lysozyme, 10 µg/ml RNase A, 10 µg/ml DNase I and 1× Complete Mini, EDTA-free protease inhibitor) as described earlier.
The cleared protein extract was incubated with 2-ml bed volume Nickel Sepharose (six Fast Flow, GE-Healthcare) at 4°C and gentle agitation. After a 30-min incubation, the Nickel Sepharose was placed into a 0.8 × 4 cm chromatography column (Poly-Prep, Biorad) and washed with 30 ml each of lysis and wash buffer (50 mM Na2HPO4 pH 7.4, 300 mM NaCl, 20 mM imidazole, 1 ml/min flow rate). The protein was eluted with 4-ml elution buffer (50 mM Na2HPO4, 300 mM NaCl, 250 mM imidazol). The eluate was placed into a dialysis tube (Spectra/Por, MWCO 12-14000) and dialyzed against 360 ml of 5 mM HEPES-KOH pH 7.4, 150 mM NaCl at 4°C with four changes of the buffer.
In vitro phosphorylation of Prp1
For in vitro phosphorylation of Prp1, 2 µg of recombinant His6-Prp1 were mixed with 1 µg Prp4 kinase in 20 µl buffer containing 20 mM HEPES-KOH pH 7.4, 3 mM MgCl2, 5% (v/v) glycerol, 0.5 mM DTT, 5 µCi γ[32P]ATP and 100 µM ATP for 30 min at 30°C. The reactions were stopped by the addition of 2× SDS sample buffer and boiling in a water bath. Proteins were separated by SDS-PAGE and autoradiography.
Phosphopeptide analysis of Prp1
Phosphopeptide analysis was performed after a protocol published by Van der Geer et al. (43). In brief, in vitro phosphorylated Prp1 was separated by SDS-PAGE and transferred onto nitrocellulose. The phosphorylated band was excised with a scalpel and incubated in 100 mM acetic acid and 0.5% PVP at 37°C for 30 min. After removal of the supernatant, the membrane was rinsed with ddH2O and freshly prepared 50 mM (NH4)2CO3. Then, the membrane was incubated with 150 µl of 50 mM (NH4)2CO3, containing 0.1 mg/ml chymotrypsin. After 2 h of incubation, another 15 µl of chymotrypsin (1 mg/ml) were added and incubation was continued for 2 h. After addition of 300 µl ddH2O, the samples were centrifuged for 5 min at 13 000 r.p.m. in a tabletop centrifuge. The supernatant was transferred into a new tube and dried in a vacuum concentrator at 4°C. The dried peptides were dissolved in 50 µl of a formic acid and 30% hydrogen peroxide mixture (9 : 1 ratio) and oxidized on ice for 1 h. After addition of 400 µl ddH2O, the samples were dried as earlier. The peptides were rinsed in 300 µl ddH2O and dried once again. The dried peptides were dissolved in 7.5 µl pH 1.9 buffer, containing 50 ml of 88% formic acid, 156 ml 100% acetic acid and 1794 ml ddH2O. Then, aliquots of 0.5 µl were spotted onto a TLC plate (20 × 20-cm glass-backed, 0.25 mm cellulose without fluorescent indicator, CEL-300 Macherey-Nagel) and dried with a fan. The plate was moistened with pH 1.9 buffer and the peptides separated for 40 min with 1000 V at 16°C. After electrophoresis, the peptides were separated in the second dimension with buffer containing 750 ml n-butanol, 500 ml pyridine, 150 ml 100% acetic acid and 600 ml ddH2O. The phosphorylated peptides were detected by autoradiography.
RESULTS
Expression of N-terminal deletions in Prp1 leads to growth arrest
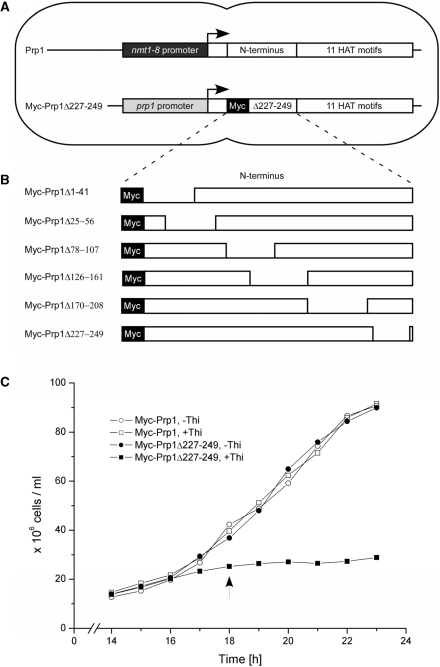
We had strong indications that Prp1 is phosphorylated in the N-terminal region at threonine residues. The N-terminus contains 17 threonines within 258 amino acids upstream of the HAT domains [http://www.digibib.tu-bs.de/?docid=00001688, (44)]. Therefore, we started a mutation analysis deleting short regions of the N-terminus containing threonines and expressed these mutations in vivo to study their consequences (Figures 1B and 6A). To be able to express solely the mutated version of Prp1 we constructed strains containing two gene copies of prp1. One copy is driven by its natural promoter and fused with its coding region to a Myc-epitope tag; the second copy is under the control of the thiamine repressible promoter nmt1-8 (Figure 1A). The control strain expresses two wild-type (wt) prp1 genes in medium without thiamine (–Thi). In medium with thiamine (+Thi), however, the nmt1-8 driven gene is repressed. This strain grows in both media with the same growth rate (Figure 1C, open circles and open squares). The deletion mutations (ΔN) were fused to the Myc-epitope tag and are driven by the natural promoter, which is constitutively transcribed. In these strains, the wt prp1 is driven by the thiamine repressible promoter. All six mutant strains show in medium –Thi the same growth rate as the control strain. However, in medium +Thi, when wt prp1 is repressed and cells express solely Prp1ΔN, all six strains arrested growth as shown here for the strain expressing Myc-Prp1Δ227-249 (Figure 1C, –Thi. filled circles; +Thi, filled squares). Under these conditions (+Thi) no wt Prp1wt protein (102 kDa) can be detected (Supplementary Figure S1A, lanes 4–6). These results reveal that the wt allele is dominant over the ΔN mutant alleles. It also suggests that structural integrity of the N-terminal region appears to be crucial for proper function of Prp1.
Figure 1.
Strains expressing N-terminal deletions of Prp1. (A) Each strain expresses a prp1+ gene controlled by the thiamine repressible promoter nmt1-8 which can be switched off by adding thiamine to the medium (+Thi). The second gene is fused in frame to an Myc-epitope-tag and driven by the natural promoter. The N-terminal mutants Myc-Prp1ΔN depicted in (B) are expressed from this gene, respectively. Both genes are integrated into the genome. (B) N-terminal deletions were generated and six strains were constructed as described (‘Materials and Methods’ section). Numbers relate to amino acid positions of Prp1 as listed in the databank. The N-terminus of Prp1 contains 258 amino acids followed by 11 HAT motifs (databank accession no. Q12381). (C) Growth behavior of strain 497 expressing Prp1 and Myc-Prp1Δ227–249 in medium without thiamine (–Thi; filled circles) and expressing solely Myc-Prp1Δ227-249 in medium with thiamine (+Thi; filled squares). Growth behavior of the control strain (470) expressing two prp1+ genes in medium without thiamine (open circles) and one prp1+ gene in medium with thiamine (open squares). In general, after 18 h protein extract was prepared from cultures with (+Thi) and without (–Thi) thiamine (arrow).
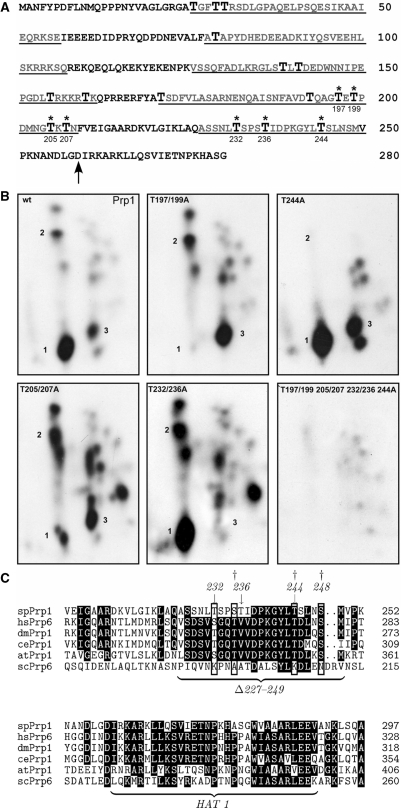
Figure 6.
In vitro and in vivo phosphorylation of Prp1. (A) Deduced amino acid sequence of the N-terminus of Prp1. Arrow indicates the beginning of the HAT1 motif (accession no. Q12381). Enlarged T, threonine; asterisks, threonines phosphorylated in vitro by Prp4 kinase. Underlined sequences are the deleted sequences as schematically shown in Figure 1B. (B) Phosphopetide maps of Prp1. In an in vitro kinase assay recombinant Prp1 was incubated with recombinant Prp4 kinase and γ[32P] ATP. After the reaction, Prp1 was separated from Prp4 and digested with chymotrypsin. The peptides were separated in two dimensions on TLC plates (‘Materials and Methods’ section). The phosphopeptides were visualized by exposure to X-ray film for 3 days. Panel wt: wild type, Prp1wt; panel: T197/195A, Prp1 contains an alanine instead of a threonine at positions 197 and 195; panel: T244A, T244A Prp1 contains an alanine in position 244; panel: T205/207A, T205/207A Prp1 contains alanine in positions 205 and 207; panel: T232/236A, Prp1 contains an alanine in positions 232 and 236; panel: T197/199/205/207/232/236/244A, Prp1 contains an alanine in positions 197/199/205/207/232/236 and 244. (C) Alignment of the HAT1 motif and the proximal N-terminal region in Prp1 with sequences of other model organisms. Clustal W alignment of the deduced S. pombe (sp) Prp1 protein (Q12381) with deduced proteins from Homo sapiens (hsPrp6, O94906); Drosophila melanogaster (dmPrp1, Q9VVU6); Caenorhabditis elegans (CePrp1, Q9GRZ2); Arabidopsis thaliana (atPrp1, Q9ZT71) and S. cerevisiae (ScPrp6, P19735). Numbers in line indicate the position of the amino acid in the protein of the respective organism as listed in the databank. Phosphorylated amino acids serines and threonines at conserved positions are boxed. Arrows indicate in vitro phosphorylation sites of Prp4 kinase determined above. Dagger at position 235 indicates a phosphorylated serine determined in phosphoproteomic analysis of fission yeast proteins (Q12381). Daggers at position 235, 244 and 248 indicate phosphorylated amino acids determined in phosphoproteomic analysis of human proteins at positions 266, 275 and 279 of hsPrp6 (O94906).
Expression of N-terminal deletions of Prp1 causes accumulation of pre-catalytic spliceosomes
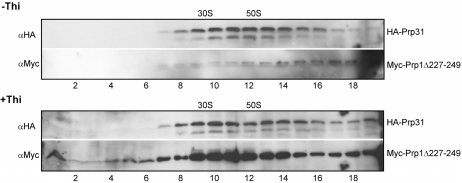
As discussed in the ‘Introduction’ section, we found Prp1 and Prp31 associated with pre-catalytic spliceosomal complexes co-sedimenting in a glycerol gradient in the range of 30–60S. Therefore, we compared the sedimentation profiles of Prp31 and Myc-Prp1Δ227–249 in a glycerol gradient when both prp1 alleles were expressed (–Thi) with the sedimentation profiles of Prp31 and Myc-Prp1Δ227–249 when cells expressed solely the mutant Myc-Prp1Δ227–249 (+Thi). This deletion is proximal to the first HAT (HAT1) motif starting from amino acid position 258 (Figure 6A, arrow). Cell extracts were isolated from growing (–Thi) and arrested cells (+Thi; Figure 1C, arrow). The gradient profile of Prp31 from growing cells expressing both Prp1wt and Myc-Prp1Δ227–249 shows, as expected, Prp31 sedimenting in the range of 30–60S (Figure 2, Panel: –Thi, HA-Prp31). Unexpectedly, probing with anti-Myc-antibodies (αMyc) to detect Myc-Prp1Δ227–249 indicates that hardly any mutant protein is sedimenting in this range with Prp31 suggesting that not much of Myc-Prp1Δ227–249 associates with these pre-catalytic spliceosomal particles. Curiously, however, full-length mutant protein Myc-Prp1Δ227–249 is not found sedimenting lower than 30S (Figure 2, Panel: −Thi, Myc-Prp1Δ227–249). By contrast, using extract of cells expressing solely Myc-Prp1Δ227–249 (+Thi), the gradient profiles show that Prp31 and mutant Prp1Δ227–249 are co-sedimenting in the range of 30–60S and some full-length mutant protein Myc-Prp1Δ227–249 is sedimenting below 30S. This suggests that the mutant protein Prp1Δ227–249 mainly binds to Prp31 complexes, if there is no wt Prp1 available (Figure 2, Panel: +Thi, HA-Prp31; and Panel: +Thi, Myc-Prp1Δ227–249). Careful analysis revealed that full-length Myc-Prp1Δ227–249, which does not associate with spliceosomal particles, is degraded mainly to cleavage products of 45 kDa sedimenting below 30S (Supplementary Figure S1A and B). To confirm this observation, we also expressed C-terminal deletions of HAT domains in this system and studied their consequences. The deletion of HAT11, for example, and sole expression of the mutant protein (Myc-Prp1Δ825–856) cause growth arrest as the expression of N-terminal deletions. The C-terminal mutant proteins, however, do not bind to spliceosomal complexes at all under both conditions and are found exclusively as 45-kDa cleavage products sedimenting in the gradient below 30S (Supplementary Figure S1C and D).
Figure 2.
Distribution of Prp31 and Prp1Δ227–249 in large complexes. Native extracts of the strain expressing HA-Prp31 and the N-terminal deletion Myc-Prp1Δ227–249 were prepared and separated on a 10–30% glycerol gradient (top fraction 1 to bottom fraction 19). Fractions 1–19 were separated by SDS-PAGE, immunoblotted and probed with anti-HA antibodies (αHA) and anti-Myc antibodies (αMyc) as indicated to determine in the gradient the distribution of HA-Prp31 and Myc-Prp1Δ227–249, respectively. Panels –Thi: extract was isolated after 18 h in medium without thiamine; Panels +Thi: extract was isolated after 18 h in medium with thiamine. The gradient was calibrated with small (30S) and large (50S) ribosomal subunits of Escherichia coli.
The strains expressing the other N-terminal deletions show the same remarkable allelic exclusion phenomenon as the strain expressing Prp1wt and Prp1Δ227–249. In addition, in all cases expressing solely Prp1ΔN, growth arrest correlates with the accumulation of pre-mRNAs of the genes rpl29, rps27, tbp1 containing one, two and three introns, respectively (data not shown).
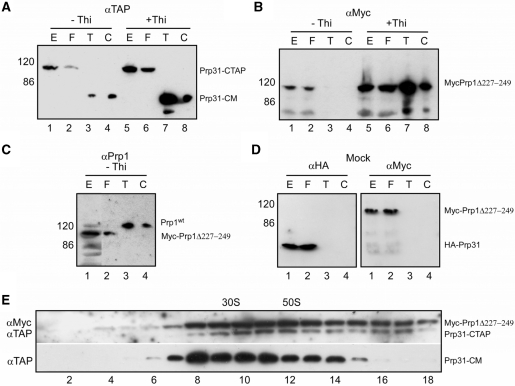
To characterize spliceosomal complexes of these strains we purified them by the tandem affinity purification (TAP) method using TAP-tagged Prp31 (26,45). Extracts of cells expressing both, Prp1wt and Myc-Prp1Δ227–249, and extracts of cells expressing solely Myc-Prp1Δ227–224 were used to purify Prp31–TAP complexes. The material eluted from the first (T) and second (C) column, respectively, was separated on SDS-PAGE, immunoblotted and probed with antibodies against the calmodulin binding peptide (αTAP) to detect Prp31. Prp31 eluted from the columns after proteolytic cleavage of the protein A region revealed the expected size of 63 kDa (Figure 3A, panel –Thi: lanes 3 and 4; panel +Thi: lanes 7 and 8: Prp31-CM). Probing the western blots with anti-Myc antibodies (αMyc) shows that purified Prp31 from growing cells is not associated with Myc-Prp1Δ227–249 (Figure 3B, panel –Thi: lanes 3 and 4), whereas in arrested cells, purified Prp31 is associated with Myc-Prp1Δ227–249 (Figure 3B, panel +Thi: lanes 7 and 8). However, probing the western blot with antibodies against Prp1 (αPrp1) shows that purified complexes of Prp31 from growing cells are exclusively associated with Prp1wt (Figure 3C, panel –Thi: lanes 3 and 4; Supplementary Figure S1A). This is consistent with the interpretation of the results received from the gradient analysis presented earlier. The sedimentation profiles of Prp31 and Prp1Δ227–249 indicated that Prp1Δ227–249 associates only with Prp31 complexes sedimenting in the range of 30–60S, if no Prp1wt is available in the cell (Figure 2, panel +Thi). After affinity purification the sedimentation profile of Prp31 from arrested cells shows that the Prp31 complexes still sediment in the range of 30–60S as observed with whole-cell extract (Figure 3E, panel: αTAP, Prp31-CM).
Figure 3.
Purification of Prp31-CTAP. Cell extracts of 5-l cultures were made from a strain expressing Prp31-CTAP and Myc-Prp1Δ227–249 after 18 h in medium without (–Thi) and with (+Thi) thiamine, respectively. Protein extract (E, 0.02% of total), flow through (F; material not binding to the first affinity column), eluate from the first affinity column IgG-Sepharose after incubation with TEV protease (T, 5% of the eluate) and eluate from the calmodulin resin after the second affinity step (C, 5% of the eluate) was separated with SDS-PAGE, immunoblotted and probed with the indicated antibodies. (A) Blot probed with anti-TAP antibodies: αTAP to detect Prp31-CTAP and Prp31-CM. (B) Blot probed with anti-Myc antibodies: αMyc to detect MycPrp1Δ227–249. (C) Blot probed with anti-Prp1 antibodies: αPrp1 to detect Prp1wt (wild type) and MycPrp1Δ227–249. (D) Mock, cell extracts were made from a strain expressing HA-Prp31 and Myc-Prp1Δ227–249 after 18 h in medium +Thi and applied consecutively to both columns as described above. Blot probed with anti-HA antibodies: αHA to detect HA-Prp31; and probed with anti-Myc antibodies: αMyc to detect Myc-Prp1Δ227-249. (E) panel αTAP: distribution of Prp31-CM. The eluate from the first affinity column (T) from extract after 18 h in medium +Thi was separated on a 10–30% glycerol gradient (top fraction 1 to bottom fraction 18). Fractions 1–18 were separated by SDS-PAGE and probed with anti-TAP (αTAP) antibodies. Panel αMyc/αTAP: extract after 18 h in medium +Thi was separated on a glycerol gradient followed by SDS-PAGE and probed with anti-Myc (αMyc) and anti-TAP (αTAP) antibodies to determine the distribution of Myc-Prp1Δ227–249 and Prp31-CTAP, respectively. The gradient was calibrated with small (30S) and large (50S) ribosomal subunits of Escherichia coli.
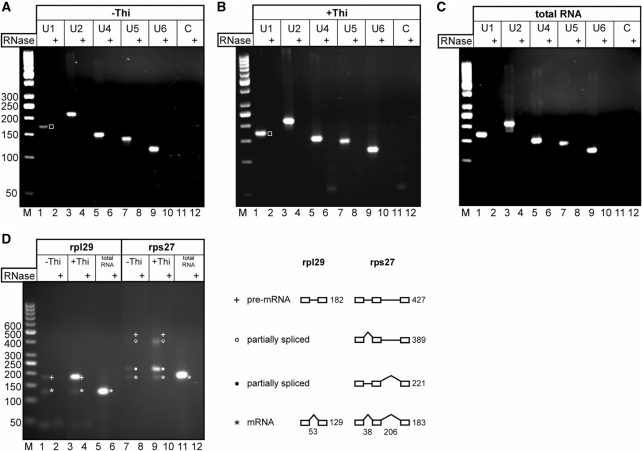
The material eluted from the second affinity step was also used to isolate RNA. We used RT-PCR to verify the presence of the U snRNAs in both samples isolated from growing (−Thi) and arrested cultures (+Thi). The results of semiquantitative RT-PCR demonstrate that the TAP purified Prp31 complexes associated with Prp1Δ227–249 in arrested cells (+Thi) contain significantly more U1 snRNA than the Prp31 complexes associated with Prp1wt from growing cells (Figure 4A and B, lane 1, open square).
Figure 4.
RT-PCR analysis to detect snRNA and pre-mRNA in affinity purified Prp31 complexes. (A, B) RNA was extracted from both eluates (±Thi) of the calmodulin resin (Figure3A, ±Thi, lane C) and subsequently analyzed by RT-PCR to determine its snRNA content. Relative levels of the five snRNAs were determined by using specific primer pairs amplifying U1, U2, U4, U5 and U6 snRNA as indicated on the top of each panel. The RT-PCR product of snRNA U1 is marked with an open square (lane 1). (C) Total RNA prepared from a wild-type strain L972 was used as a positive control. (D) The same RNA samples as in (A) and (B) were analyzed by RT-PCR to determine their (pre)-mRNA content. Specific primer pairs were used to amplify (pre)-mRNA of the ribosomal protein genes rpl29 and rps27 as indicated. The exon/intron structure of both genes is shown to the right. Numbers below indicate intron length in nucleotides for both pre-mRNAs. Numbers to the right indicate expected RT-PCR product sizes in base pairs. All RNA samples were treated with DNase I to remove possible DNA contaminants prior to reverse transcription. Complete removal of contaminating DNA was verified by RNase A treatment of the RNA as indicated (RNase+). RT–PCR products were separated in 3% agarose gels and stained with ethidiumbromide. Lanes marked with ‘C’: negative control without template using the U1 primer pair. Lanes M: marker (50-bp ladder); numbers to the left indicate fragment length in base pairs.
The RNA isolated from the spliceosomal Prp31 complexes after the second affinity step was also used to probe for the presence of pre-mRNAs and mature mRNAs of the ribosomal protein genes rpl29 and rps27, respectively. The gene rpl29 contains a single intron of 53 nt in size, whereas rps27 contains two introns of 38 and 206 nt in size, respectively. The results of semiquantitative RT-PCR using total RNA of logarithmically growing cells show that both ribosomal protein genes are expressed and, particularly, effectively spliced showing no trace amount of unspliced pre-mRNA (Figure 4D, lanes 5 and 11). In arrested cultures (+Thi) where the Prp31 complexes are associated with Prp1Δ227–249, the results of semiquantitative RT-PCR show that the TAP purified Prp31 complexes contain significant amounts of unspliced rpl29 pre-mRNA and only trace amounts of mature mRNA (Figure 4D, lane 3), whereas Prp31 complexes isolated from growing cells (−Thi) contain only trace amounts of pre-mRNA and mRNA (Figure 4D, lane 1). In the case of rps27, the Prp31 complexes isolated from arrested cultures (+Thi) accumulate partially spliced and spliced pre-mRNAs (Figure 4D, lanes 7 and 9).
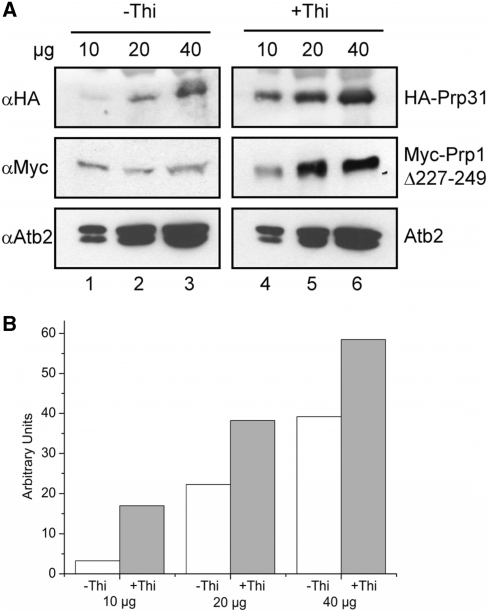
Taken together, the observations described above prompted us to suggest that the sole expression of Prp1Δ227–249 leads to stalled pre-catalytic spliceosomes containing pre-mRNAs and consisting of the five U snRNPs. Thus, we may expect accumulation of Prp31 in cells solely expressing Prp1Δ227–249. Therefore, we quantitated the relative amount of Prp31 in extracts of growing cells (−Thi) and compared it with the relative amount of Prp31 measured in extracts of arrested cells (+Thi). The results revealed that the amount of Prp31 is 2–4-fold increased in arrested cells, indicating that pre-catalytic spliceosomal particles associated with Prp31 and Prp1Δ227–249 accumulate in these cells (Figure 5A and B).
Figure 5.
The amount of Prp31 associated with large pre-catalytic particles increases in cells expressing solely Myc-Prp1Δ227–249. (A) Protein extract from cells cultured 18 hours in medium without (–Thi) and with (+Thi) thiamine, respectively, was separated by SDS-PAGE, immunoblotted and probed with anti-HA antibodies (αHA), anti-Myc antibodies (αMyc) and anti-Atb2 antibodies (αAtb2) as indicated. αHA visualizes HA-Prp31, αMyc visualizes Myc-Prp1Δ227–249 and αAtb2 visualizes Atb2 (α-tubulin). To each lane 10, 20, 40-µg protein extract was applied, respectively. (B) Blots were analyzed using the Bio-Rad GelDoc XR/ChemiDoc XRS gel documentation system and protein band densities were analyzed using the Quantity One software provided with the image Station. White columns relative amount of HA-Prp31 in cells cultured in medium −Thi; gray columns relative amount of HA-Prp31 in cells cultured in medium +Thi; 10, 20 and 40 µg protein was used as indicated in (A).
To confirm that Prp31 is associated with native pre-catalytic spliceosomes containing the five snRNAs, including basepaired U4/U6, we immunoprecipitated extracts of cells expressing HA-Prp31 with HA-antibodies (αHA) and characterized the snRNA composition of the immunoprecipitates using a solution hybridization assay that preserved base pairing of U4 and U6 (26,42). When Prp31 was immunoprecipitated from extracts of growing (−Thi) and arrested (+Thi) cultures, the spliceosomal complexes always contained the five snRNAs including base paired U4/U6 (Supplementary Figure S2). This result and the fact that Prp31 always sediments in a relatively broad range of 30–60S confirm that spliceosomal particles containing five snRNAs U1, U2, U5, U4/U6 exist in vivo in a pool of pre-catalytic particles of different size. The data presented are consistent with the notion that expression of Prp1Δ227–249 leads to the accumulation of pre-catalytic spliceosomal particles containing the five snRNAs and pre-mRNA, indicating that manipulation in the N-terminal region of Prp1 prevents activation, but not assembly of spliceosomes.
Phosphorylation sites of Prp4 kinase in the N-terminus of Prp1
Prp1 is a physiological substrate of Prp4 kinase; therefore, we wanted to determine the phosphorylation sites in Prp1. Prp4 kinase phosphorylates Prp1 in vitro at threonine residues in the N-terminal domain (44). The N-terminus of Prp1 contains 17 threonine residues distributed throughout the 258 amino acids as indicated (Figure 6A). Since no phosphorylation site sequences of Prp4 kinase were known, we changed the 17 threonine residues in the N-terminus by site-specific mutagenesis to alanine and produced the mutated Prp1 in bacteria. Recombinant Prp1 and Prp4 kinase were incubated in a kinase assay with γ[32P]ATP followed by phosphopeptide analysis of Prp1. The phosphopeptide map of Prp1wt revealed three strongly labeled spots and several weak signals (Figure 6B, panel: wt Prp1, spots 1–3). When we used a Prp1 protein in which the threonine at position 244 was changed to an alanine (T244A-Prp1), for example, spot 2 disappears in the peptide map, demonstrating that position 244 was phosphorylated (Figure 6B, panel: T244A). The results of the mapping analyses show that positions T197/199, T205/207, T232/236 and T244 are unequivocally in vitro phosphorylation sites of Prp4 kinase in the N-terminus of Prp1 (Figure 6B). These sites are located proximal to the first HAT domain, whereas T232, T236 and T244 reside in the region which has been deleted in Myc-Prp1Δ227–249 and T197/199, T205/207 reside in the region deleted in Myc-Prp1Δ170-208 (Figures 6A and 1B).
The alignment of the amino acid sequences comprising 50 amino acids of the N-terminus proximal to the first HAT motif (HAT1) of Prp1 with human Prp6, and with orthologs of genetic model organisms, including S. cerevisiae, shows that the phosphorylation sites in this region (amino acids 227–249) are conserved in all organisms except in the N-terminus of Prp6 of S. cerevisiae (Figure 6C). It is noteworthy here that in phosphoproteome analysis of fission yeast a phosphopeptide was detected which carried a modified serine at position 235 [Figure 6C, dagger, (46)]. Furthermore, even more intriguingly, phosphoproteome analysis of human cells revealed peptides phosphorylated at position T235, T244 and S248, respectively (Figure 6C; daggers, hsPrp6, O94906).
DISCUSSION
Consequences of the expression of N-terminal mutations in Prp1: accumulation of spliceosomes containing U1, U2, U5 and U4/U6
In this study, we investigated the function of the N-terminus of the S. pombe spliceosomal protein Prp1, which is the ortholog of S. cerevisiae and human Prp6 (U5-102 K). This is the first report demonstrating that the N-terminal region of Prp1 is essential to mediate a splicing event; however, it is not necessary for assembly of spliceosomes in vivo. The 11 HAT domains comprising the C-terminus of Prp1 mediate interaction with other spliceosomal components. Several genetic interactions, including extragenic suppressors and synthetic lethal interactions with prp1, have been found. These studies yielded temperature-sensitive (ts) prp1 alleles containing mutations in different HAT motifs and established an epistasis map showing, in addition to Prp4 kinase, genetic interactions with prp13 (encoding snRNA U4 in S. pombe, snu4) and other spliceosomal components, such as Prp6 (Snu13 Sc, 15,5K Hs), Prp5 (Prp46 Sc, PRL1 Hs), Spp101 (Lin1/Snu40 Sc, U5-55K Hs) and Spp42 (Prp8 Sc, U5-220K Hs), which is involved in coordinating activities of several splicing factors at the heart of the spliceosome (12–14,17,47–51 and Razanau and Käufer, unpublished data). However, none of these studies revealed a ts allele of prp1 containing a mutation in the N-terminal region of the protein.
Therefore, we expressed N-terminal mutations in prp1 in a genetic background which contained a repressible wt gene. Cells expressing both Prp1wt and Prp1Δ227–249 grow with the same growth rate as cells expressing two wt genes (Figure 1C). Under these conditions, almost no mutant Prp1 associates with the spliceosomal particles sedimenting between 30 and 60 S. Although our data presented in this report reveal that mutant Prp1 does not interfere with the function of Prp1wt and that mutant Prp1 not associated with spliceosomal particles eventually becomes degraded, we have no data explaining this phenomenon of extreme allelic exclusion. Therefore, we are prone to speculate. The observation that the wt Prp1wt completely outcompetes the mutant protein is consistent with the idea that Prp1 is involved in the activation of spliceosomes. Assuming that pre-mRNA splicing activity is high in growing cells, one would expect that the level of splicing competent spliceosomes containing Prp1wt and all five snRNPs is very low. We have made many observations which indicate that Prp1 plays a special role in regulating pre-mRNA splicing in fission yeast. For example, after depletion of Prp1 by switching off the prp1 gene, pre-mRNA accumulates and pre-spliceosomal complexes containing Prp31 and all five snRNAs still exist, although Prp1wt protein was not detectable anymore (14).
In this report, we showed that in cells solely expressing Prp1Δ227–249 the increasing amounts of Prp31 and U1 snRNA correlate with the accumulation of unspliced pre-mRNA associated with pre-catalytic spliceosomes. This is an indication that the expression of Prp1Δ227–249 does not prevent assembly of spliceosomes on pre-mRNA. We do not know, and cannot probe at this point, whether a proper or aberrant pre-mRNA splicing complex is assembled when Prp1Δ227–249 is expressed. However, our data are consistent with the notion that expression of Prp1Δ227–249 causes spliceosomes to stall before activation. Interestingly, all the N-terminal mutations investigated show similar phenotypes, emphasizing that the N-terminus of Prp1 is crucial for activation of spliceosomes in vivo.
We determined the phosphorylation sites of Prp4 kinase in Prp1. The generation of the phosphopeptide maps revealed that seven threonine residues in the N-terminus of Prp1 are strongly phosphorylated by Prp4 kinase in vitro (Figure 6A, asterisks). Our experimental approach to determine by in vivo labeling and subsequent phosphopeptide mapping whether the in vitro sites are also used in vivo was cumbersome and revealed convincing data only for the threonine residue at position 244 (Figure 6C and data not shown). However, phospho-proteomic analyses of fission yeast and of human cell extracts identified phosphates at the amino acid residues in positions 235, 244 and 248, indicating that these amino acids are phosphorylated in vivo. Together, these findings led us suggest that this region is a target of kinases, including Prp4. Most intriguingly, this region, including the positions of the phosphorylation sites, is highly conserved throughout the kingdoms with the remarkable exception of Prp6 of S. cerevisiae (including all Hemiascomycetes, Figure 6C).
We still do not know what the molecular consequences are, when Prp1 is phosphorylated by Prp4 kinase in vivo. However, based on our results presented here and before, we hypothesize that in fission yeast phosphorylation of Prp1 by Prp4 kinase is part of the process in which spliceosomes are activated (13).
In an elegant study demonstrating that rem1 encoding a meiotic cyclin is only expressed when a 87-nt intron is spliced out, it has been shown that pre-mRNA splicing is coupled with transcription in fission yeast. Splicing of rem1-pre-mRNA is dependent on the meiosis specific transcription factor Mei4 regulating transcription of the rem1 gene and recruiting the splicing machinery to the transcribed locus (52–54). This is one of the very few examples of pre-mRNA splicing regulation in fission yeast so far. However, in fission yeast about 45% of the genes contain introns. Genes contain from one to more than 10 introns and the introns are small with a mean size of 78 nt (55). Thus, we reason that in general removal of introns in fission yeast is constitutive by default and, if splicing is coupled with transcription the spliceosomes need to be activated at transcribed loci after the splicing apparatus has recognized the introns to be spliced out. We suggest that phosphorylation of Prp1 by Prp4 kinase plays a role within the described scenario—either the phosphorylation by Prp4 kinase is part of a mechanism that signals that an intron is occupied by a splicing competent spliceosome in a sense of quality control and/or the phosphorylation is directly involved in inducing the rearrangements for catalysis. In any case, our hypothesis can be experimentally approached and further analysis will help to provide insight into the complex control mechanisms exerted to express intron-containing genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Aleh Razanau is a scholar of the Georg-Christoph-Lichtenberg program “Molecular Complexes of Biomedical Relevance” at the Technische Universität Braunschweig working towards a PhD. The government of the federal state Niedersachsen is funding the Georg-Christoph-Lichtenberg programs. Funding to pay the Open Access publication charge was provided by Technische Universität Braunschweig.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Sperling J, Azubel M, Sperling R. Structure and function of the pre-mRNA splicing machine. Structure. 2008;16:1605–1615. doi: 10.1016/j.str.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Brow DA. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 5.Rino J, Carmo-Fonseca M. The spliceosome: a self-organized macromolecular machine in the nucleus? Trends Cell. Biol. 2009;19:375–384. doi: 10.1016/j.tcb.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Görnemann J, Kotvic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–64. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA: 5′ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Tardif DF, Lacadie SA, Rosbash M. A genome wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol. Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 10.Nilsen TW. Spliceosome assembly in yeast: one Chip at a time? Nat. Struct. Mol. Biol. 2005;12:571–573. doi: 10.1038/nsmb0705-571. [DOI] [PubMed] [Google Scholar]
- 11.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 12.Schwelnus W, Richert K, Opitz F, Groß T, Habara Y, Tani T, Käufer NF. Fission yeast Prp4p kinase regulates pre-mRNA splicing by phosphorylating a non-SR-splicing factor. EMBO Rep. 2001;2:35–41. doi: 10.1093/embo-reports/kve009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn AN, Käufer NF. Pre-mRNA splicing in Schizosaccharomyces pombe; regulatory role of a kinase conserved from fission yeast to mammals. Curr. Gene. 2003;42:241–251. doi: 10.1007/s00294-002-0355-2. [DOI] [PubMed] [Google Scholar]
- 14.Bottner CA, Schmidt H, Vogel S, Michele M, Käufer NF. Multiple genetic and biochemical interactions of Brr2, Prp8, Prp31, Prp1p and Prp4 kinase suggest a function in the control of the activation of spliceosomes in Schizosaccharomyces pombe. Curr. Genet. 2005;48:151–161. doi: 10.1007/s00294-005-0013-6. [DOI] [PubMed] [Google Scholar]
- 15.Dellaire G, Makarov EM, Cowger J, Longman D, Sutherland HGE, Lührmann R, Torchia J, Bickmore WA. Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol. Cell. Biol. 2002;22:5141–5156. doi: 10.1128/MCB.22.14.5141-5156.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YIG, Moore RE, Ge HY, Young MK, Lee TD, Stevens SW. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35:3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urushiyama S, Tani T, Ohshima Y. The prp1 gene required for pre-mRNA splicing in Schizosaccharomyces pombe encodes a protein that contains TPR motifs and is similar to Prp6p of budding yeast. Genetics. 1997;147:101–115. doi: 10.1093/genetics/147.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bon E, Casaregola S, Blandin G, Llorente B, Neuveglise C, Munsterkotter M, Guldener U, Mewes HW, Van Helden J, Dujon B, et al. Molecular evolution of eukaryotic genomes: hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 2003;31:1121–1135. doi: 10.1093/nar/gkg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Häcker I, Sander B, Golas MM, Wolf E, Karagöz E, Kastner B, Stark H, Fabrizio P, Lührman R. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat. Struct. Mol. Biol. 2009;15:1207–1212. doi: 10.1038/nsmb.1506. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Li P, Dybkov O, Nottrott S, Hartmuth G, Lührmann R, Carlomagno T, Wahl M. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2008;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 21.Schaffert N, Hossbach M, Heintzmann R, Achsel T, Lührmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm A, Lührmann R. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 2004;23:2381–2391. doi: 10.1038/sj.emboj.7600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarov EM, Makarova OV, Achsel T, Lührmann R. The human homologue of the yeast splicing factor Prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein–protein interactions. J. Mol. Biol. 2000;298:567–575. doi: 10.1006/jmbi.2000.3685. [DOI] [PubMed] [Google Scholar]
- 24.Galisson F, Legrain P. The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the (U4/U6.U5) tri-snRNP. Nucleic Acids Res. 1993;21:1555–1562. doi: 10.1093/nar/21.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang T, Vilardell J, Query CC. Pre-spliceosome formation in S. pombe requires a stable complex of SF1-U2AF(59)-U2AF(23) EMBO J. 2002;21:5516–5526. doi: 10.1093/emboj/cdf555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newo AN, Lützelberger M, Bottner CA, Wehland J, Wissing J, Jänsch L, Käufer NF. Proteomic analysis of the U1 snRNP of Schizosaccharomyces pombe reveals three essential organism-specific proteins. Nucleic Acids Res. 2007;35:1391–1401. doi: 10.1093/nar/gkl1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald WH, Ohi R, Smelkova N, Frendewey D, Gould KL. Myb-related fission yeast Cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 1999;19:5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohi MD, Vander Kooi CW, Rosenberg JA, Ren L, Hirsch JP, Chazin WJ, Walz T, Gould KL. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol. Cell. Biol. 2005;25:451–460. doi: 10.1128/MCB.25.1.451-460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohi MD, Ren L, Wall JS, Gould KL, Walz T. Structural chracterization of the fission yeast U5.U2/U6 spliceosomal complex. Proc. Natl Sci. USA. 2007;104:3195–3200. doi: 10.1073/pnas.0611591104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carnahan RH, Feoktistova A, Ren L, Niessen S, Yates JR, III, Gould KL. Dim1p is required for efficient splicing and export of mRNA encoding Lid1p, a component of the fission yeast anaphase-promoting complex. Eukaryotic Cell. 2005;4:577–587. doi: 10.1128/EC.4.3.577-587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King RC, editor. Handbook of Genetics. Vol. 1. New York: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 33.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 34.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 35.Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- 37.Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 38.Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- 39.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 40.Gould KL, Ren L, Feoktistova AS, Jennings JL, Link AJ. Tandem affinity purification and identification of protein complex components. Methods. 2004;33:239–244. doi: 10.1016/j.ymeth.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 41.Berger SL, Birkenmeier CS. Inhibition of intractable nucleases with ribonucleoside – vanadyl complexes: isolation of messenger ribonucleic acid from resting lymphocytes. Biochemistry. 1979;18:5143–5149. doi: 10.1021/bi00590a018. [DOI] [PubMed] [Google Scholar]
- 42.Li Z, Brow DA. A rapid assay for quantitative detection of specific RNAs. Nucleic Acids Res. 1993;21:4645–4646. doi: 10.1093/nar/21.19.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Geer P, Luo K, Sefthorn BM, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis on cellulose thin-layer plates. In: Hardie DG, editor. Protein Phosphorylation – A Practical Approach. New York: Oxford University Press; 1993. p. 97. [Google Scholar]
- 44.Schwelnus W. Dissertation. Germany: Technical University of Braunschweig; 2004. [Google Scholar]
- 45.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotech. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 46.Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J. Proteome Res. 2008;7:1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 47.Potashkin J, Li R, Frendewey D. Pre-mRNA splicing mutants of Schizosaccharomyces pombe. EMBO J. 1989;8:551–559. doi: 10.1002/j.1460-2075.1989.tb03409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenberg GH, Alahari SK, Käufer NF. prp4 from Schizosaccharomyces pombe, a mutant deficient in pre-mRNA splicing isolated using genes containing artificial introns. Mol. Gen. Genet. 1991;226:305–309. doi: 10.1007/BF00273617. [DOI] [PubMed] [Google Scholar]
- 49.Urushiyama S, Tani T, Oshima Y. Isolation of novel pre-mRNA mutants of Schizosaccharomyces pombe. Mol. Gen. Genet. 1996;253:118–127. doi: 10.1007/s004380050304. [DOI] [PubMed] [Google Scholar]
- 50.Gross T, Lützelberger M, Weigmann H, Klingenhoff A, Shenoy S, Käufer NF. Functional analysis of the fission yeast Prp4 protein kinase involved in pre-mRNA splicing and isolation of a putative mammalian homologue. Nucleic Acids Res. 1997;25:1028–1035. doi: 10.1093/nar/25.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Potashkin J, Kim D, Fons M, Humphrey T, Frendewey D. Cell-division-cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr. Genet. 1998;34:153–163. doi: 10.1007/s002940050381. [DOI] [PubMed] [Google Scholar]
- 52.Malpeira J, Moldon A, Hidalgo E, Smith GR, Nurse P, Ayte J. A meiosis-specific cyclin regulated by splicing is required for proper progression through meiosis. Mol. Cell. Biol. 2005;25:6330–6337. doi: 10.1128/MCB.25.15.6330-6337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moldon A, Malpeira J, Gabrielli N, Gogol M, Gomez-Escoda B, Ivanova T, Seidel C, Ayte J. Promoter-driven splicing regulation in fission yeast. Nature. 2008;455:997–1000. doi: 10.1038/nature07325. [DOI] [PubMed] [Google Scholar]
- 54.Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol. Cell. 2005;18:491–498. doi: 10.1016/j.molcel.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 55.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.