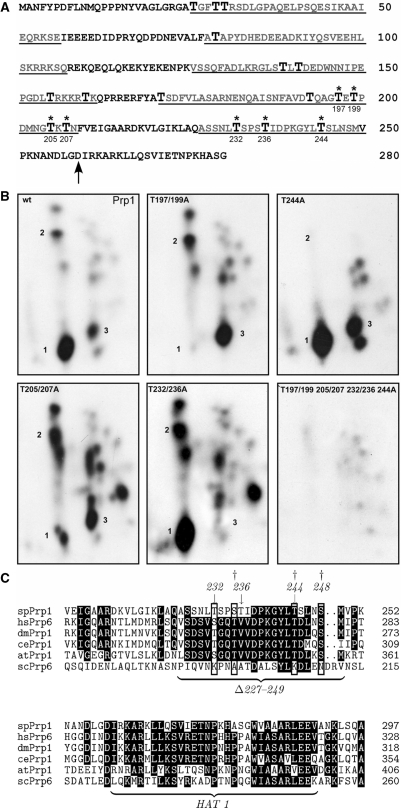
Figure 6.
In vitro and in vivo phosphorylation of Prp1. (A) Deduced amino acid sequence of the N-terminus of Prp1. Arrow indicates the beginning of the HAT1 motif (accession no. Q12381). Enlarged T, threonine; asterisks, threonines phosphorylated in vitro by Prp4 kinase. Underlined sequences are the deleted sequences as schematically shown in Figure 1B. (B) Phosphopetide maps of Prp1. In an in vitro kinase assay recombinant Prp1 was incubated with recombinant Prp4 kinase and γ[32P] ATP. After the reaction, Prp1 was separated from Prp4 and digested with chymotrypsin. The peptides were separated in two dimensions on TLC plates (‘Materials and Methods’ section). The phosphopeptides were visualized by exposure to X-ray film for 3 days. Panel wt: wild type, Prp1wt; panel: T197/195A, Prp1 contains an alanine instead of a threonine at positions 197 and 195; panel: T244A, T244A Prp1 contains an alanine in position 244; panel: T205/207A, T205/207A Prp1 contains alanine in positions 205 and 207; panel: T232/236A, Prp1 contains an alanine in positions 232 and 236; panel: T197/199/205/207/232/236/244A, Prp1 contains an alanine in positions 197/199/205/207/232/236 and 244. (C) Alignment of the HAT1 motif and the proximal N-terminal region in Prp1 with sequences of other model organisms. Clustal W alignment of the deduced S. pombe (sp) Prp1 protein (Q12381) with deduced proteins from Homo sapiens (hsPrp6, O94906); Drosophila melanogaster (dmPrp1, Q9VVU6); Caenorhabditis elegans (CePrp1, Q9GRZ2); Arabidopsis thaliana (atPrp1, Q9ZT71) and S. cerevisiae (ScPrp6, P19735). Numbers in line indicate the position of the amino acid in the protein of the respective organism as listed in the databank. Phosphorylated amino acids serines and threonines at conserved positions are boxed. Arrows indicate in vitro phosphorylation sites of Prp4 kinase determined above. Dagger at position 235 indicates a phosphorylated serine determined in phosphoproteomic analysis of fission yeast proteins (Q12381). Daggers at position 235, 244 and 248 indicate phosphorylated amino acids determined in phosphoproteomic analysis of human proteins at positions 266, 275 and 279 of hsPrp6 (O94906).