Abstract
The catalytic importance of enzyme active-site interactions is frequently assessed by mutating specific residues and measuring the resulting rate reductions. This approach has been used in bacterial ketosteroid isomerase to probe the energetic importance of active-site hydrogen bonds donated to the dienolate reaction intermediate. The conservative Tyr16Phe mutation impairs catalysis by 105-fold, far larger than the effects of hydrogen bond mutations in other enzymes. However, the less-conservative Tyr16Ser mutation, which also perturbs the Tyr16 hydrogen bond, results in a less-severe 102-fold rate reduction. To understand the paradoxical effects of these mutations and clarify the energetic importance of the Tyr16 hydrogen bond, we have determined the 1.6-Å resolution x-ray structure of the intermediate analogue, equilenin, bound to the Tyr16Ser mutant and measured the rate effects of mutating Tyr16 to Ser, Thr, Ala, and Gly. The nearly identical 200-fold rate reductions of these mutations, together with the 6.4-Å distance observed between the Ser16 hydroxyl and equilenin oxygens in the x-ray structure, strongly suggest that the more moderate rate effect of this mutant is not due to maintenance of a hydrogen bond from Ser at position 16. These results, additional spectroscopic observations, and prior structural studies suggest that the Tyr16Phe mutation results in unfavorable interactions with the dienolate intermediate beyond loss of a hydrogen bond, thereby exaggerating the apparent energetic benefit of the Tyr16 hydrogen bond relative to the solution reaction. These results underscore the complex energetics of hydrogen bonding interactions and site-directed mutagenesis experiments.
Keywords: active-site environment, enzymatic catalysis, protein cavities, site-directed mutagenesis
Our knowledge of enzyme function has been greatly advanced by a combination of structure determination and site-directed mutagenesis. These approaches have yielded detailed models of active-site architectures and identified the key catalytic groups positioned near bound substrates whose mutation results in large rate reductions. From this knowledge and an understanding of basic properties of chemical reactivity, detailed reaction mechanisms have been elucidated for numerous enzymes (1). Nevertheless, incisive understanding of the energetic contributions of physical interactions within active sites to the extraordinary 1010–1020-fold rate enhancements of enzymes remains a central challenge of biochemistry and a key hurdle in rational enzyme design (2–6).
Site-directed mutagenesis has been used extensively in bacterial ketosteroid isomerase from Pseudomonas putida (pKSI) and Commamonas testosteroni (tKSI) to probe the catalytic importance of active-site hydrogen bonds. KSI catalyzes a reversible double-bond isomerization within steroid substrates that proceeds via a dienolate intermediate stabilized by hydrogen bonds donated by Tyr16 (pKSI numbering) and protonated Asp103 within an active-site oxyanion hole (Fig. 1A). The conservative Tyr16Phe mutation decreases kcat by approximately 50,000-fold (see additional discussion in SI Text), corresponding to a 6.3 kcal/mol reduction in catalysis that is far larger than the effects of hydrogen bond mutations in other enzymes (Table S1) (7, 8). The large rate reduction from this mutation, along with observation of a far-downfield NMR chemical shift for the Tyr16 hydroxyl proton and a short 2.5-Å O⋯O distance between Tyr16 and the intermediate analogue equilenin, led to suggestions that the Tyr16 hydrogen bond donated to the reacting substrate was energetically more favorable than hydrogen bonds formed to water molecules in the nonenzymatic solution reaction and contributed approximately 6 kcal/mol to specific transition state stabilization by KSI (9–12).
Fig. 1.
KSI substrates and reaction intermediate analogue. (A) Reaction mechanism for isomerization of 5(10)-estrene-3,17-dione (5(10)-EST). (B) Isomerization of 5-androstene-3,17-dione (5-AND). (C) Schematic depiction of equilenin bound at the KSI active site.
This interpretation of the unusually large rate reduction of the Tyr16Phe mutation suggested that any mutation that ablated the hydrogen bond donated by Tyr16 to the reacting substrate would give rise to a similarly large 6 kcal/mol reduction in catalysis. Paradoxically, the less-conservative Tyr16Ser mutation was reported to result in a much less severe 30-fold reduction in kcat, despite the simple expectation that this mutation would also disrupt the Tyr16 hydrogen bond (13). The authors accounted for this result by suggesting that the Tyr16Ser mutant was still capable of forming a hydrogen bond to the dienolate intermediate. Direct formation of a hydrogen bond between the Ser16 hydroxyl and the intermediate would require an approximately 5-Å restructuring of the KSI backbone in the vicinity of residue 16 to position a Ser within hydrogen-bonding distance of the bound substrate. Alternatively, other active-site changes mediated by the Tyr16Ser mutation could permit hydrogen bond formation by other groups at this position.
To clarify the paradoxical effects of the Tyr16Phe and Tyr16Ser mutations, to test the model that a Ser at position 16 can donate a hydrogen bond to the dienolate intermediate, and to better understand the catalytic contributions of active-site hydrogen bonds, we have determined the 1.6-Å resolution x-ray structure of the intermediate analogue, equilenin, bound to the Tyr16Ser mutant and tested the requirement for a hydroxyl group at position 16 by mutating Tyr16 to Ser, Thr, Ala, and Gly. The nearly identical 3 kcal/mol rate reductions of these mutations, together with the 6.4-Å distance observed between the Ser16 hydroxyl and equilenin oxygen atoms in the x-ray structure, provide direct evidence that the more moderate rate effect of this mutant is not due to maintenance of a hydrogen bond from Ser at position 16. These results, additional spectroscopic observations, and prior structural studies suggest an alternative model to explain the unusually large rate reduction of the Tyr16Phe mutation and the more moderate effect of the Tyr16Ser mutant and provide a cautionary lesson for interpreting hydrogen bond energetics from site-directed mutagenesis experiments.
Results and Discussion
Rate Effects from Mutating Tyr16 to Phe, Ser, Thr, Ala, and Gly.
Prior measurements of the Tyr16Phe and Tyr16Ser rate effects were performed using a steroid substrate, 5-androstene-3,17-dione (5-AND, Fig. 1B), for which chemical steps are not cleanly rate-limiting (14). Thus, the reported rate reductions may represent lower limits for the effects of these mutations on chemical catalysis of steroid isomerization. To clarify the rate effects on active-site chemistry in the absence of contributions from substrate binding or product release, we determined kcat and KM for wild-type and mutant pKSI using a substrate, 5(10)-estrene-3,17-dione [5(10)-EST, Fig. 1A], for which chemical steps are cleanly rate-limiting (15). Our results with 5(10)-EST are summarized in Fig. 2 and Table S2 and are identical, within error, to independently determined values we reported in a separate study (16). The Tyr16Phe mutation decreases kcat by 20,000-fold, corresponding to a 5.8 kcal/mol reduction in catalysis. This result is very similar to the previously reported kcat reduction of 50,000-fold (6.3 kcal/mol energetic effect) determined for tKSI Tyr16Phe using 5-AND (7). With 5(10)-EST, the Tyr16Ser mutation reduces kcat by 300-fold, a 10-fold larger decrease than previously determined using 5-AND (13) but a 100-fold smaller value than the effect from the Tyr16Phe mutation.
Fig. 2.
Effects of Tyr16 mutations on KSI activity (kcat). Values and errors are averages and standard deviations from three or more independent measurements at different enzyme concentrations and are from Table S2. The indicated values for the Tyr16Ser and Tyr16Phe mutants are identical within error to values we recently reported in a separate publication (16).
Why does the less-conservative Tyr16Ser mutation result in a smaller rate decrease than the conservative Tyr16Phe mutant? A prior study proposed that the Ser16 hydroxyl maintained a hydrogen bond to the dienolate reaction intermediate that was ablated in the Phe16 mutant (13). To test this proposal, we measured the rate reductions upon mutation of Tyr16 to additional groups that either preserved (Thr) or ablated (Ala, Gly) the hydrogen-bonding ability of residue 16. If the presence or absence of a hydrogen bond to a mutated residue 16 resulted in the moderate (approximately 300-fold) and large (approximately 20,000-fold) rate reduction previously observed for the Ser and Phe mutants, respectively, then we expected to observe a moderate rate reduction for the Thr mutant but a much larger decrease for the Ala and Gly mutants at this position. We observed an approximately 200-fold kcat reduction for all three mutants. This reduction is nearly identical to the approximately 300-fold Tyr16Ser decrease (Fig. 2), strongly suggesting that the moderate energetic effect of the Ser16 mutation does not arise from formation of a hydrogen bond between residue 16 and the dienolate intermediate.
To further test the prior model and clarify the physical origin of the paradoxical rate effects of Tyr16 mutations, we determined the x-ray structure of the Tyr16Ser mutant and compared it to existing structures for wild-type and Tyr16Phe KSI.
X-Ray Structure of the Intermediate Analogue, Equilenin, Bound to Tyr16Ser KSI and Comparison to Wild-Type and Tyr16Phe KSI.
We determined the x-ray structure of the intermediate analogue, equilenin (Fig. 1C), bound to pKSI Tyr16Ser/Asp40Asn.* X-ray data collection and model refinement statistics are listed in Table S3 (Rwork = 20.7% and Rfree = 24.5%). The overall KSI structure obtained at 1.6-Å resolution is nearly identical to that observed previously in a 1.1-Å resolution structure of equilenin bound to wild-type pKSI [Protein Data Bank (PDB) code 1OH0], with a root-mean-square deviation between the two structures of 0.4 Å for backbone atoms (10). The electron density map for the Tyr16Ser structure, contoured in Fig. 3A at 1.5σ, shows well-defined densities for the modeled atomic positions within the oxyanion hole, with no indication of alternative ligand or side-chain orientations.
Fig. 3.
Structural studies of KSI. (A) Sigma-A-weighted 2Fo - Fc electron density map (contoured at 1.5σ) from the 1.6-Å resolution structure of equilenin bound to Tyr16Ser/Asp40Asn pKSI. Distances are average values (standard deviation ± 0.1 Å) from the four independently refined monomers contained in the asymmetric unit. (B) Superposition of the pKSI Tyr16Ser/Asp40Asn·equilenin structure determined herein (carbon atoms colored cyan), the 1.8-Å resolution structure of unliganded pKSI Tyr16Phe (PDB entry 1EA2, carbon atoms colored violet), and the 1.1-Å resolution structure of equilenin bound to wild-type pKSI (PDB entry 1OH0, carbon atoms colored green). (C) Triangle-shaped sigma-A-weighted 2Fo - Fc electron density (cyan mesh, contoured at 1.5σ) assigned to partially disordered water within the approximately 100 Å3 cavity resulting from the Tyr16Ser mutation. Yellow dashes connect groups within hydrogen-bonding distance of visible electron density to the region of greatest electron density (visible up to 3.3σ). (D) Downfield region of 1H NMR spectrum of equilenin bound to tKSI Tyr16Phe/Asp40Asn. The peak at approximately 13 ppm is present in spectra of unliganded KSI (11, 25).
The refined orientation of bound equilenin is nearly identical to that observed previously for the pKSI·equilenin complex, with equilenin positioned to accept a short hydrogen bond from the similarly positioned side chain of Asp103 (O⋯O distance ∼2.5 Å) in both structures (Fig. 3B). The hydrogen bond between Tyr16 and equilenin, however, has been ablated by the Tyr16Ser mutation. The backbone atoms of Ser16 are positioned nearly identically to those of Tyr16 in wild-type KSI, and the 6.4-Å distance observed between the Ser16 hydroxyl and equilenin oxygens precludes formation of a hydrogen bond between these groups. Rather, the Ser16 side chain is positioned to donate a hydrogen bond (O⋯O distance 2.7 Å) to the backbone carbonyl oxygen of Met13 (Fig. 3A). No other enzymatic group is positioned within hydrogen-bonding distance (3.5 Å) of the equilenin oxygen. On the basis of these observations and the kinetic results described above, we conclude that the moderate rate reduction of the Tyr16Ser mutation is not due to maintenance of a hydrogen bond between Ser16 and the dienolate intermediate, nor does the Ser hydroxyl aid catalysis relative to other similarly sized side chains that lack a hydrogen bond donor.
An alternative explanation is suggested by additional observations from the Tyr16Ser structure, as follows. The KSI active site is readily accessed in its unliganded state by water molecules from bulk solvent. Removal of the Tyr16 phenol moiety via mutation to Ser generates a cavity of approximately 100 Å3 within the KSI oxyanion hole (17) that is not filled in by structural rearrangements of nearby groups. This volume is sufficient to accommodate three to four water molecules that can remain within the cavity upon ligand binding. While the cavity surface has substantial hydrophobic character imparted by the aliphatic Met, Ile, and Val side chains that formerly packed around the hydrophobic phenyl ring of Tyr16, the apical cavity regions are polar and bracketed at one end by Ser16 and the other end by Asp103, Tyr57, Tyr32, and the oxygen of bound equilenin (Fig. 3C). These polar groups may hydrogen bond with adventitious water molecules and thus promote their retention within the cavity. Indeed, prior structural studies have suggested that two to three polar contacts within a protein cavity are sufficient to retain water (18–20).
Consistent with the presence of several partially disordered water molecules that occupy multiple near-in-space conformations within the cavity created by the Tyr16Ser mutation, we observed triangle-shaped electron density in the 2Fo - Fc electron density map proximal to Tyr57, Tyr32, and the equilenin oxygen (Fig. 3C). This density could not be accounted for by modeling any of the small molecules present in the crystallization or purification buffers into the refined structure and was best described by two to three water molecules with occupancies split between multiple positions (see additional discussion in SI Methods). The region of greatest electron density (visible up to 3.3σ) is located 3.0 Å and 2.7 Å from the oxygens of equilenin and Tyr57, respectively, suggesting substantial occupancy of a water conformation that is within hydrogen-bonding distance of these two groups. Additional support for the presence of water within the Tyr16Ser cavity is provided by 19F NMR experiments described in the following section.
On the basis of these observations, we propose that the moderate rate decrease of the Tyr16Ser mutation is due to the presence of several water molecules within the oxyanion hole cavity that solvate the reacting substrate via hydrogen bond formation and thus mitigate the catalytic consequences of ablating the hydrogen bond from Tyr16. This model is consistent with the nearly identical rate effects of the Tyr16 to Ser, Thr, Ala, and Gly mutations, all of which are expected to result in similar-sized cavities that retain water. Furthermore, the distance between Ser16 and the region of greatest electron density in the cavity is more than 4.3 Å, suggesting minimal interactions between residue 16 and cavity water and consistent with the nearly identical rate effects of the above mutants.
If the moderate rate effect of the less-conservative Tyr16Ser mutation can be explained on the basis of forming an active site cavity containing water molecules that solvate the reacting substrate, what physical interactions underpin the large rate reduction of the more conservative Tyr16Phe mutation? Previous studies have attributed the unusually large rate decrease of this mutation to loss of an exceptionally strong hydrogen bond to the dienolate intermediate (9–11, 21). What are the structural consequences of the Tyr16Phe mutation, and does this mutation lead to additional stabilizing or destabilizing interactions beyond loss of a hydrogen bond?
Two x-ray structures have been reported for pKSI containing the Tyr16Phe mutation: a 1.8-Å resolution structure of apo Tyr16Phe (PDB code 1EA2) (21) and a 1.9-Å resolution structure of Tyr16Phe/Asp40Asn with bound equilenin (PDB code 1OHO) (22). In the latter structure, equilenin was reported to bind in a backward conformation with the carbonyl oxygen of the distal D-ring of the steroid positioned within the oxyanion hole. On the basis of this observation, the authors suggested that nonproductive substrate binding to Tyr16Phe KSI might contribute to the large kcat reduction of this mutation (13). However, nonproductive substrate binding is expected to decrease both kcat and KM to a similar extent (1). Our observation of only a twofold KM difference between wild-type and Tyr16Phe pKSI (Table S2) strongly suggests that nonproductive substrate binding does not contribute significantly to the large rate reduction of the Tyr16Phe mutant. The reported backward binding of equilenin to Tyr16Phe/Asp40Asn may therefore be unique to the Asp40Asn-containing double mutant or a result of the particular crystallization conditions used that led to extensive protein precipitation (22).
To identify possible interactions between the dienolate reaction intermediate and the Tyr16Phe active site that might clarify the physical basis of its strongly attenuated catalysis, we generated a structural model of the Tyr16Phe mutant with equilenin bound in the productive, canonical conformation by superposition of the published 1.8-Å resolution Tyr16Phe apo structure with the 1.1-Å resolution wild-type pKSI·equilenin structure (Fig. 3B). While the Tyr16Phe mutation disrupts the Tyr16 hydrogen bond, our structural model suggests that Asp103 and the equilenin oxygen remain within hydrogen-bonding distance. Support for formation of this hydrogen bond is provided by our observation of a far-downfield resonance at 17.4 ppm in the 1H NMR spectrum of equilenin bound to tKSI Tyr16Phe/Asp40Asn (Fig. 3D), as expected for removal of one but not both of the short hydrogen bonds observed in the 1.1-Å resolution x-ray structure of equilenin bound to wild-type pKSI.† Our structural model suggests that no group other than Asp103 donates a hydrogen bond within the oxyanion hole of the Tyr16Phe mutant.
Does the Tyr16Phe mutation create space for a water molecule, as observed for the Tyr1Ser mutant? As shown in Fig. 3B, the phenyl ring of Phe16 is slightly repositioned by approximately 1 Å relative to Tyr16, as previously reported (21). A similar repositioning of the Phe16 ring is observed in the 1.9-Å resolution Tyr16Phe/Asp40Asn structure (PDB entry 1OHO). While the Phe mutation and this structural rearrangement liberate approximately 20 Å3 formerly occupied by the ring and oxygen atoms of Tyr16, this volume is distributed along the entire length of the Phe16 ring and is thus insufficiently condensed to accommodate a water molecule. Indeed, neither of the Tyr16Phe structures reported a water molecule within this liberated space proximal to Phe16.‡ The available structures therefore suggest that the Tyr16Phe mutation disrupts the Tyr16 hydrogen bond and retains the phenyl ring of Phe16 within close proximity (approximately 3.5 Å) of the substrate oxygen, preventing formation of a water-occupied cavity and any attendant stabilizing interactions with the reaction intermediate. This close approximation of a hydrophobic phenyl ring to the substrate oxygen on which negative charge localizes along the reaction coordinate (Fig. 1A) may introduce additional destabilizing interactions that exacerbate the energetic consequence of the Tyr16Phe mutation beyond loss of an enzymatic hydrogen bond and replacement by a solution hydrogen bond.
Does mutation of Tyr16 to Phe result in an apolar local solvation environment within the oxyanion hole near residue 16 while mutation to Ser retains water-like solvation in this region, as suggested by our structural analysis and the above-stated model? To directly assess differences in the local solvation environment as a consequence of the Tyr16Ser or Tyr16Phe mutations, we turned to 19F NMR studies of the intermediate analogue, 2-fluoro-4-nitrophenolate, bound to KSI.
Probing the Local Solvation Environment within the Oxyanion Hole of Tyr16Ser and Tyr16Phe KSI via 19F NMR.
The 19F NMR chemical shift of a fluorine atom provides a sensitive probe of its surrounding chemical environment (23). Thus, a fluorine atom positioned within the KSI oxyanion hole in the vicinity of residue 16 would report on changes in the local solvation environment introduced by mutation of Tyr16 to Phe or Ser.
Single-ring phenolates bind to the KSI active site and serve as analogues of the dienolate reaction intermediate (24, 25). A prior structural study of 2-fluorophenolate bound to pKSI Asp40Asn indicated that this ligand populated a single conformation in the oxyanion hole, with the ortho-F atom exclusively oriented toward Tyr16 (26). This orientation appeared to be enforced by unfavorable repulsive interactions that occurred if the ligand adopted an alternative conformation with the fluorine atom positioned proximal to Asp103. The side chain of Asp103 is nearly identically oriented in x-ray structures of Tyr16Ser, Tyr16Phe, and wild-type pKSI (Fig. 3B). Based on this observation, a phenolate containing a 2-fluoro group is expected to bind both mutants with the ortho-F atom positioned near residue 16 (to avoid unfavorable interactions with Asp103) and thus provide a probe of the local solvation environment in this region of the mutated oxyanion holes (Fig. 4A).
Fig. 4.
Binding of 2-fluorophenolate reaction intermediate analogues to pKSI. (A) Structural model of 2-fluorophenolate (carbon atoms in yellow, fluorine in black) bound to pKSI Tyr16Ser (carbon atoms and electron density of disordered water in Ser16 cavity colored cyan) and pKSI Tyr16Phe (carbon atoms colored violet, Phe16 shown as space-filling) generated by superposition of pKSI Asp40Asn·2 - fluorophenolate (PDB entry 3CPO), pKSI Tyr16Ser/Asp40Asn·equilenin (PDB entry 3IPT), and pKSI Tyr16Phe (PDB entry 1EA2). (B) 19F NMR spectra of 2-fluoro-4-nitrophenolate (pKa = 6.0) in aqueous solution (pH 12), in the aprotic and low polarity organic solvent THF containing excess triethylamine to form the phenolate anion, and bound to pKSI Tyr16Ser/Asp40Asn or pKSI Tyr16Phe/Asp40Asn.
We acquired 19F NMR spectra of 2-fluoro-4-nitrophenolate (pKa = 6.0) bound to pKSI Tyr16Ser/Asp40Asn and Tyr16Phe/Asp40Asn. UV-vis spectra confirmed binding to these mutants as the ionized phenolate (Fig. S1). As shown in Fig. 4B, the 19F nucleus resonates with a substantially different chemical shift when bound to the two mutants, with the -134.7 ppm peak in the Phe16 mutant shifted nearly 2 ppm downfield from the -136.4 ppm peak detected for the Ser16 mutant.
We first considered whether ring current effects from the phenyl ring of Phe16 that are absent in the Ser16 mutant could account for the observed chemical shift difference. Structural modeling of 2-fluorophenolate bound to Tyr16Phe (Fig. 4A) suggests that the ortho-F atom is approximately 4.5 Å from the phenyl ring of Phe16. Previous ab initio and density functional theory calculations of 19F NMR shielding contributions from a benzene ring positioned near hexafluorobenzene, however, reported negligible shielding contributions at separation distances greater than 4.0 Å (27). Thus, ring current effects from Phe16 are not expected to contribute significantly to chemical shift differences of the fluorine nucleus in Tyr16Phe versus Tyr16Ser.
The observed 2 ppm dispersion in the chemical shift of the ortho-F nucleus positioned proximal to Phe16 or the Ser16 cavity therefore suggests substantial differences in the local solvation environment in this region of the oxyanion hole. To calibrate the change in 19F NMR chemical shift expected for a change in solvation environment, we acquired spectra of the 2-fluoro-4-nitrophenolate in water and in the aprotic, low polarity solvent tetrahydrofuran (THF). As shown in Fig. 4B, transfer of the phenolate from water into THF deshields the fluorine nucleus by 2.2 ppm, very similar to the change we observe for KSI. Furthermore, the observed chemical shift in THF is nearly identical to the value observed for the phenolate bound to Tyr16Phe, while the detected peak in aqueous solution is within 0.5 ppm of the peak position in Tyr16Ser. These observations support the model based on our structural and energetic results that the Tyr16Ser mutation replaces Tyr16 with a water-occupied cavity while the Tyr16Phe mutation replaces Tyr16 with a desolvating and hydrophobic local environment.
Conclusions and Implications.
Site-directed mutagenesis has provided a facile means to disrupt specific active-site interactions and assess the degree to which this disruption impairs enzyme activity. Whereas it has been tempting to interpret a rate decrease upon mutation as a measurement of the specific energetic contribution of an individual residue or interaction to catalysis, the physical and energetic connection between wild-type and mutant enzymes is complex (1, 28–31). Enzymes accelerate reactions relative to their nonenzymatic rates in neutral water (2). Evaluation of the catalytic contribution of specific physical interactions within an enzyme active site must therefore specify the energetic benefit of an enzymatic interaction relative to corresponding interactions formed during the water-solvated nonenzymatic reaction. In the case of enzymatic hydrogen bonds donated to a reacting substrate, energetic comparison should be made to hydrogen bonds donated by water to the reacting substrate and not simply to the absence of a hydrogen bond (1).
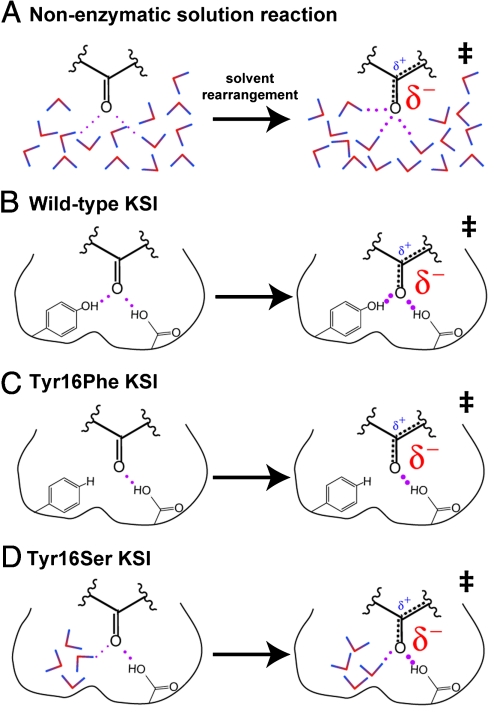
Multiple prior studies have suggested that the large (approximately 6 kcal/mol) rate reduction observed for the Tyr16Phe mutation in the KSI oxyanion hole, together with unusual physical properties associated with the Tyr16 hydrogen bond, provides evidence for such a large catalytic contribution from hydrogen bond formation by Tyr16 during steroid isomerization relative to the nonenzymatic reaction in water (Fig. 5A and B) (9–11). Our results and analysis, however, suggest that the Phe mutation ablates the hydrogen bond donated by Tyr16 and replaces it with a hydrophobic surface within the oxyanion hole (Fig. 5C). These changes collectively desolvate the dienolate intermediate and thereby exaggerate the apparent energetic benefit of the Tyr16 hydrogen bond relative to water.
Fig. 5.
Schematic models for the effects of Tyr16 mutations on KSI catalysis. Shorter hydrogen bonds are depicted with thicker dots. (A) Solution nonenzymatic reaction with water molecules shown as colored dipoles that reorient to solvate localized charge in the dienolate-like transition state. (B) The wild-type oxyanion hole has two positioned enzyme groups that donate hydrogen bonds to the reacting substrate that may strengthen in the transition state. (C) The Tyr16Phe mutation ablates one of the oxyanion hole hydrogen bonds but does not permit water entry, resulting in a hydrophobic surface that desolvates the localized negative charge in the dienolate-like transition state. (D) The Tyr16Ser mutation replaces Tyr16 with a water-filled cavity that provides aqueous-like solvation of the reacting substrate.
Mutation of Tyr16 to the much smaller residues of Ser, Thr, Ala, or Gly results in a moderate rate reduction of approximately 200-fold that is nearly identical for all four mutants and similar to the 100- to 1000-fold activity decreases observed for oxyanion hole mutations in other enzymes (Table S1). Our results strongly suggest that the moderate rate effects from these mutants are due to creation of a water-filled cavity that functionally replaces the Tyr16 hydrogen bond with aqueous-like solvation of the dienolate intermediate (Fig. 5D).
The observed 200-fold rate reductions of these mutants may therefore provide a crude estimate of the catalytic contribution from the Tyr16 hydrogen bond relative to hydrogen bonds formed to water in the nonenzymatic reference reaction. But we emphasize the crudeness of this estimate. Its accuracy requires that the energetics of interactions with water in the newly formed cavity be similar to those with water in the water-solvated solution reaction and further requires that additional structural rearrangements do not propagate from the Tyr16Ser mutation. Two structural observations suggest that these conditions may be generally satisfied. First, the positions of equilenin and Asp103 observed for the Tyr16Ser mutant were nearly identical to those observed previously in the wild-type pKSI·equilenin complex (Fig. 2B), suggesting minimal transmission of structural changes distal to the site of mutation. Indeed, the nearby Ser16 cavity and the presence of several waters did not significantly lengthen the observed hydrogen bond O⋯O distance between Asp103 and equilenin, consistent with a previous study that suggested a tightly packed environment within the KSI active site and strong constraints on the position of Asp103 (26). Second, water within the Ser16 cavity is sufficiently ordered to observe electron density by x-ray diffraction yet sufficiently disordered such that we were not able to refine the observed electron density into discrete water sites. This partial ordering due to confinement within the cavity and hydrogen-bonding contacts with several nearby polar groups (Fig. 3C) may crudely approximate the diminished mobility and partial ordering of water molecules observed for the primary hydration shell around hydrophobic solutes and charged hydrogen bond acceptor groups (32–35), both features of the reacting steroid substrate of KSI.
Nevertheless, solvation and hydrogen bonding are highly complex. The apparent approximately 200-fold catalytic contribution of the Tyr16 hydrogen bond relative to water is the net result of multiple possible favorable and unfavorable energetic contributions, as is the case with all active-site hydrogen bonds (see additional discussion in SI Text). The individual energetic components of hydrogen bond contributions to catalysis can, in principle, be most incisively dissected by computation, although experimental studies represent a reality that cannot be matched in silico. Multiple experimental techniques and comparisons will provide the richest datasets for interpretation, and nontrivial computational predictions of new experimental behaviors will provide a necessary and powerful guide for future experiments and computation that lead toward a deep and complete understanding of hydrogen bond energetics and enzymatic catalysis.
Methods
A full description of all experimental methods is given in SI Text. Plasmids encoding KSI Tyr16 mutants were created by Quik-Change (Stratagene) site-directed mutagenesis and confirmed by DNA sequencing. Mutant and wild-type enzymes were expressed and purified from Escherichia coli using published methods (16, 25), and their kinetic constants for isomerization of 5(10)-EST were determined as previously described (16). Blade-shaped cocrystals of pKSI Tyr16Ser/Asp40Asn containing bound equilenin in space group P21 were obtained at 20 °C using hanging drop vapor diffusion by mixing 2 μL of 30 mg/ml KSI containing 1 molar equivalent of equilenin with 2 μL of reservoir solution (0.9 M ammonium sulfate, 40 mM potassium phosphate, pH 7). Diffraction data at 100 K were collected at beamline 8.2.2 of the Advanced Light Source (Lawrence Berkeley National Laboratory). Data refinement was carried out as previously described (25, 26) with minor modifications, and statistics for data collection and refinement are summarized in Table S3. Structure figures were prepared using MacPyMOL (36). 19F NMR spectra of 2-fluoro-4-nitrophenolate were acquired at 20 °C on a 500-MHz Varian UNITYINOVA NMR spectrometer as previously described (26). 1H NMR spectra of 300 μM equilenin bound to 300 μM tKSI Tyr16Phe/Asp40Asn were acquired at -3 °C on an 800-MHz Varian UNITYINOVA NMR spectrometer equipped with a 5-mm, triple-resonance, gradient 1H (13C/15N) probe using previously published methods (26).
Supplementary Material
Acknowledgments.
This work was funded by a National Science Foundation grant to D.H. (MCB-0641393). D.A.K. and P.A.S. were supported in part by Howard Hughes Medical Institute Predoctoral Fellowships. P.A.S received additional support from a G. Lieberman Fellowship. T.D.F. was supported by the University-wide AIDS Research Program of the University of California (F03-ST-216). Portions of this research were conducted at the Advanced Light Source, a national user facility operated by the Lawrence Berkeley National Laboratory. We thank Axel Brunger for support and helpful discussions and members of the Herschlag lab for comments on the manuscript.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors for equilenin bound to the pKSI mutant Tyr16Ser/Asp40Asn have been deposited in the Protein Data Bank with accession code 3IPT.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911168107/DCSupplemental.
*The Asp40Asn mutation mimics the protonated general base, Asp40, present in the KSI-intermediate complex.
†A prior NMR study of equilenin bound to tKSI Tyr16Phe/Asp40Asn performed on a 600-MHz instrument reported that no downfield peaks above 14 ppm were observed (37). On a higher field and more sensitive 800-MHz instrument, we indeed observe a far-downfield peak at 17.4 ppm for this complex, as expected for a short hydrogen bond between Asp103 and equilenin.
‡The apparent absence of water proximal to Phe16 is consistent with our prior observation that substitution of Tyr16 with unnatural para-bromo-Phe, para-chloro-Phe, and para-methyl-Phe derivatives results in nearly identical rate reductions as the Phe mutation, as expected if no bound water is displaced by these bulky para substituents (38).
References
- 1.Fersht AR. Structure and Mechanism in Protein Science. New York: W.H. Freeman and Company; 1999. [Google Scholar]
- 2.Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 3.Jiang L, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc Natl Acad Sci USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlt JA, Babbitt PC. Enzyme (re)design: Lessons from natural evolution and computation. Curr Opin Chem Biol. 2009;13:10–18. doi: 10.1016/j.cbpa.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lad C, Williams NH, Wolfenden R. The rate of hydrolysis of phosphomonoester dianions and the exceptional catalytic proficiencies of protein and inositol phosphatases. Proc Natl Acad Sci USA. 2003;100:5607–5610. doi: 10.1073/pnas.0631607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuliopulos A, Mildvan AS, Shortle D, Talalay P. Kinetic and ultraviolet spectroscopic studies of active-site mutants of delta 5-3-ketosteroid isomerase. Biochemistry. 1989;28:149–159. doi: 10.1021/bi00427a022. [DOI] [PubMed] [Google Scholar]
- 8.Kim DH, et al. Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from pseudomonas putida biotype b. Biochemistry. 2000;39:4581–4589. doi: 10.1021/bi992119u. [DOI] [PubMed] [Google Scholar]
- 9.Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J Biol Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 10.Kim SW, et al. High-resolution crystal structures of delta 5-3-ketosteroid isomerase with and without a reaction intermediate analogue. Biochemistry. 1997;36:14030–14036. doi: 10.1021/bi971546+. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Q, Abeygunawardana C, Talalay P, Mildvan AS. NMR evidence for the participation of a low-barrier hydrogen bond in the mechanism of delta 5-3-ketosteroid isomerase. Proc Natl Acad Sci USA. 1996;93:8220–8224. doi: 10.1073/pnas.93.16.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feierberg I, Aqvist J. The catalytic power of ketosteroid isomerase investigated by computer simulation. Biochemistry. 2002;41:15728–15735. doi: 10.1021/bi026873i. [DOI] [PubMed] [Google Scholar]
- 13.Nam GH, et al. Maintenance of alpha-helical structures by phenyl rings in the active-site tyrosine triad contributes to catalysis and stability of ketosteroid isomerase from Pseudomonas putida biotype b. Biochemistry. 2001;40:13529–13537. doi: 10.1021/bi015547k. [DOI] [PubMed] [Google Scholar]
- 14.Hawkinson DC, Eames TC, Pollack RM. Energetics of 3-oxo-delta 5-steroid isomerase: Source of the catalytic power of the enzyme. Biochemistry. 1991;30:10849–10858. doi: 10.1021/bi00109a007. [DOI] [PubMed] [Google Scholar]
- 15.Pollack RM, Bantia S, Bounds PL, Koffman BM. pH dependence of the kinetic parameters for 3-oxo-delta 5-steroid isomerase. Substrate catalysis and inhibition by (3s)-spiro[5 alpha- androstane-3,2’-oxiran]-17-one. Biochemistry. 1986;25:1905–1911. doi: 10.1021/bi00356a011. [DOI] [PubMed] [Google Scholar]
- 16.Schwans JP, Kraut DA, Herschlag DA. Determining the catalytic role of remote substrate binding interactions in ketosteroid isomerase. Proc Natl Acad Sci USA. 2009;106:14271–14275. doi: 10.1073/pnas.0901032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai J, Taylor R, Chothia C, Gerstein M. The packing density in proteins: Standard radii and volumes. J Mol Biol. 1999;290:253–266. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
- 18.Williams MA, Goodfellow JM, Thornton JM. Buried waters and internal cavities in monomeric proteins. Protein Sci. 1994;3:1224–1235. doi: 10.1002/pro.5560030808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard SJ, Gross KH, Argos P. Intramolecular cavities in globular proteins. Protein Eng. 1994;7:613–626. doi: 10.1093/protein/7.5.613. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, Quillin ML, Matthews BW. Use of experimental crystallographic phases to examine the hydration of polar and nonpolar cavities in T4 lysozyme. Proc Natl Acad Sci USA. 2008;105:14406–14411. doi: 10.1073/pnas.0806307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi G, et al. Pseudoreversion of the catalytic activity of Y14F by the additional substitution(s) of tyrosine with phenylalanine in the hydrogen bond network of delta 5-3-ketosteroid isomerase from Pseudomonas putida biotype b. Biochemistry. 2001;40:6828–6835. doi: 10.1021/bi002767+. [DOI] [PubMed] [Google Scholar]
- 22.Oh KS, et al. Role of catalytic residues in enzymatic mechanisms of homologous ketosteroid isomerases. Biochemistry. 2000;39:13891–13896. doi: 10.1021/bi001629h. [DOI] [PubMed] [Google Scholar]
- 23.Gerig JT. Fluorine nuclear magnetic resonance of fluorinated ligands. Methods Enzymol. 1989;177:3–23. doi: 10.1016/0076-6879(89)77003-8. [DOI] [PubMed] [Google Scholar]
- 24.Petrounia IP, Pollack RM. Substituent effects on the binding of phenols to the D38N mutant of 3-oxo-delta5-steroid isomerase. A probe for the nature of hydrogen bonding to the intermediate. Biochemistry. 1998;37:700–705. doi: 10.1021/bi972262s. [DOI] [PubMed] [Google Scholar]
- 25.Kraut DA, et al. Testing electrostatic complementarity in enzyme catalysis: Hydrogen bonding in the ketosteroid isomerase oxyanion hole. PLoS Biol. 2006;4:501–519. doi: 10.1371/journal.pbio.0040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sigala PA, et al. Testing geometrical discrimination within an enzyme active site: Constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole. J Am Chem Soc. 2008;130:13696–13708. doi: 10.1021/ja803928m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DerHovanessian A, Rablen PR, Jain A. Ab initio and density functional calculations of 19F NMR chemical shifts for models of carbonic anhydrase inhibitors. J Phys Chem A. 2000;104:6056–6061. [Google Scholar]
- 28.Kraut DA, Carroll KS, Herschlag D. Challenges in enzyme mechanism and energetics. Annu Rev Biochem. 2003;72:517–571. doi: 10.1146/annurev.biochem.72.121801.161617. [DOI] [PubMed] [Google Scholar]
- 29.Knowles JR. Tinkering with enzymes: What are we learning? Science. 1987;236:1252–1258. doi: 10.1126/science.3296192. [DOI] [PubMed] [Google Scholar]
- 30.Fersht AR. Relationships between apparent binding energies measured in site-directed mutagenesis experiments and energetics of binding and catalysis. Biochemistry. 1988;27:1577–1580. doi: 10.1021/bi00405a027. [DOI] [PubMed] [Google Scholar]
- 31.Plapp BV. Site-directed mutagenesis: A tool for studying enzyme catalysis. Method Enzymol. 1995;249:91–119. doi: 10.1016/0076-6879(95)49032-9. [DOI] [PubMed] [Google Scholar]
- 32.Harpham MR, Levinger NE, Ladanyi BM. An investigation of water dynamics in binary mixtures of water and dimethyl sulfoxide. J Phys Chem B. 2008;112:283–293. doi: 10.1021/jp074985j. [DOI] [PubMed] [Google Scholar]
- 33.Perera PN, et al. Observation of water dangling OH bonds around dissolved nonpolar groups. Proc Natl Acad Sci USA. 2009;106:12230–12234. doi: 10.1073/pnas.0903675106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laage D, Stirnemann G, Hynes JT. Why water reorientation slows without iceberg formation around hydrophobic solutes. J Phys Chem B. 2009;113:2428–2435. doi: 10.1021/jp809521t. [DOI] [PubMed] [Google Scholar]
- 35.Timmer RL, Bakker HJ. Hydrogen bond fluctuations of the hydration shell of the bromide anion. J Phys Chem A. 2009;113:6104–6110. doi: 10.1021/jp902424u. [DOI] [PubMed] [Google Scholar]
- 36.DeLano WL. MacPyMOL: A PyMOL-based molecular graphics application for MacOS X. Palo Alto, CA: DeLano Scientific LLC; 2007. [Google Scholar]
- 37.Zhao Q, Abeygunawardana C, Gittis AG, Mildvan AS. Hydrogen bonding at the active site of delta 5-3-ketosteroid isomerase. Biochemistry. 1997;36:14616–14626. doi: 10.1021/bi971549m. [DOI] [PubMed] [Google Scholar]
- 38.Kraut DA, Churchill MJ, Dawson PE, Herschlag D. Evaluating the potential for halogen bonding in the oxyanion hole of ketosteroid isomerase using unnatural amino acid mutagenesis. ACS Chem Biol. 2009;4:269–273. doi: 10.1021/cb900016q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.