Fig. 4.
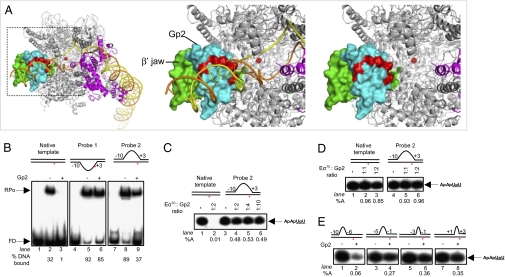
Gp2 inhibits step(s) leading to the RPo. (A) Model of the Gp2–RNAP complex. The boxed region in the image in the left panel is enlarged in the middle (with promoter DNA) and right panels (without promoter DNA) to emphasize the β′ jaw region. The RNAP is presented as in Fig. 1A. Gp2 is shown in cyan, and the negatively charged side chains of residues E21, E28, E34, D37, E38, E41, E44 and E53, which protrude into the DNA binding channel, are highlighted in red. (B) Autoradiograph of a 4.5% (wt/vol) native gel showing heparin-resistant RPo formation by Eσ70 in the absence (lanes 2, 5, and 8) and presence (lanes 3, 6, and 9) of ∼2-fold molar excess of Gp2 on 32P-labeled versions of the fully duplex (native) and heteroduplex probes 1 and 2 lacUV5 promoter templates (see text; FD, free DNA). The % template DNA in the RPo is shown at the bottom of the gel. (C) Autoradiograph of a 20% (wt/vol) denaturing gel showing synthesis of the transcript ApApUpU (indicated by the arrow) from the native (lanes 1 and 2) and heteroduplex probe 2 (lanes 3–6) lacUV5 promoter templates in the absence (lanes 1 and 3) and presence (lanes 2, 4, 5, and 6) of Gp2. The Gp2:Eσ70 molar ratio in each lane is shown at the Top. The percentage transcripts synthesized (%A) by Eσ70 in the presence of Gp2 with respect to reactions with no Gp2 are given at the bottom. (D) As in C, but Gp2 was added to the reactions after transcriptionally competent promoter complexes had formed on native (lanes 2 and 3) and heteroduplex probe 2 (lanes 5 and 6) lacUV5 promoter templates. (E) As in C, but assays were conducted with lacUV5 promoter templates containing heteroduplex segments of different lengths and at different positions with respect to the transcription start site (probes 3–6; see text). In B–E, the lacUV5 promoter templates used are shown schematically with the positions and lengths of the heteroduplex segment indicated with respect to +1 site (indicated by the red asterisk).