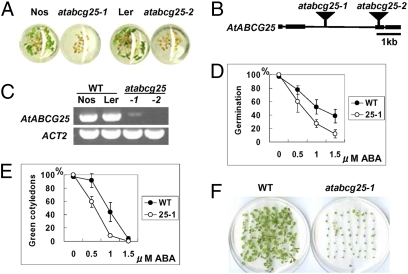
Fig. 1.
Identification of the AtABCG25 gene and atabcg25 mutant alleles. (A) Isolation of ABA-sensitive mutants by 96-well multititer plate assay. Compared with wild-types (Nos, Ler), the mutants (atabcg25-1, atabcg25-2) were more sensitive to 1.0 μM ABA solution. The titer plate was incubated in a growth chamber under long-day conditions for 7 days. (B) AtABCG25 gene structure and insertional mutation sites of two atabcg25 alleles. Square boxes represent exons, and black bars represent introns. Transposon insertions in atabcg25-1 and atabcg25-2 are shown as triangles. (C) AtABCG25 transcripts in wild-type plants and mutants identified by RT-PCR analysis. Total RNAs were prepared from two wild types (WT) and two atabcg25 mutants (atabcg25): Nossen (Nos) and Landsberg (Ler), and atabcg25-1 (-1) and atabcg25-2 (-2), respectively. Actin2 (ACT2) was used as a reference. (D–F) ABA-hypersensitive phenotype of atabcg25-1. Seed germination (D) and postgerminative growth (E) were scored for the wild type (WT) and atabcg25-1 mutant (25-1) in different concentrations of ABA at day 2 (D) and day 4 (E). Values are shown as mean ± SD of 50 seeds (obtained from three independent experiments). Photographs were taken of seedlings of wild type (WT) (F, Left) and atabcg25-1 (atabcg25-1) (F, Right) germinated in the presence of 1.0 μM ABA. Fifty seeds of each type were sown and incubated for 18 days.