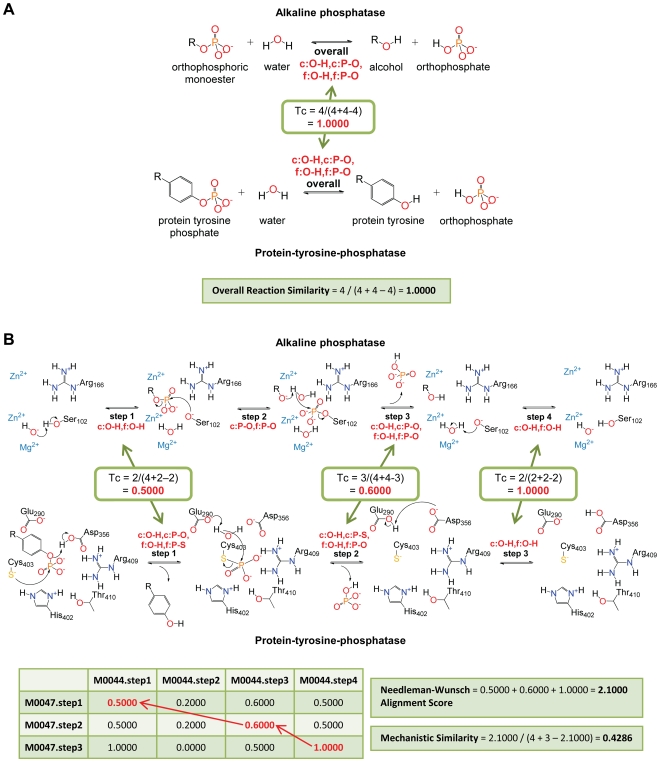
Figure 1. Quantification of overall reaction and mechanistic similarity.
The reactions catalyzed by alkaline phosphatase (MACiE M0044, EC 3.1.3.1, PDB 1alk) [118]–[120], and protein-tyrosine-phosphatase (MACiE M0047, EC 3.1.3.48, PDB 1ytw) [121]–[123] are used as examples. Each reaction in MACiE is described as an overall transformation (A) and as a sequence of mechanistic steps (B). For measuring reaction similarity, each overall reaction and mechanistic step is represented as the set of bond changes occurring in the transformation from substrates to products, with c: bond cleaved, d: bond decreased in order, f: bond formed, and i: bond increased in order. Similarity between sets of bond changes is computed using Tanimoto coefficients (Tc). (A) Overall similarity is computed as the Tanimoto coefficient between the set of bond changes occurring in the transformation of substrates to products of the reactions. (B) Mechanistic similarity is computed from a global alignment of the mechanistic steps. First, Tanimoto coefficients between all possible pairs of steps are stored in a similarity matrix, and then the maximum-match pathway is obtained using the Needleman-Wunsch algorithm. To obtain the mechanistic similarity a new Tanimoto coefficient is computed using the number of steps in each reaction and the Needleman-Wunsch alignment score as inputs (see Methods).