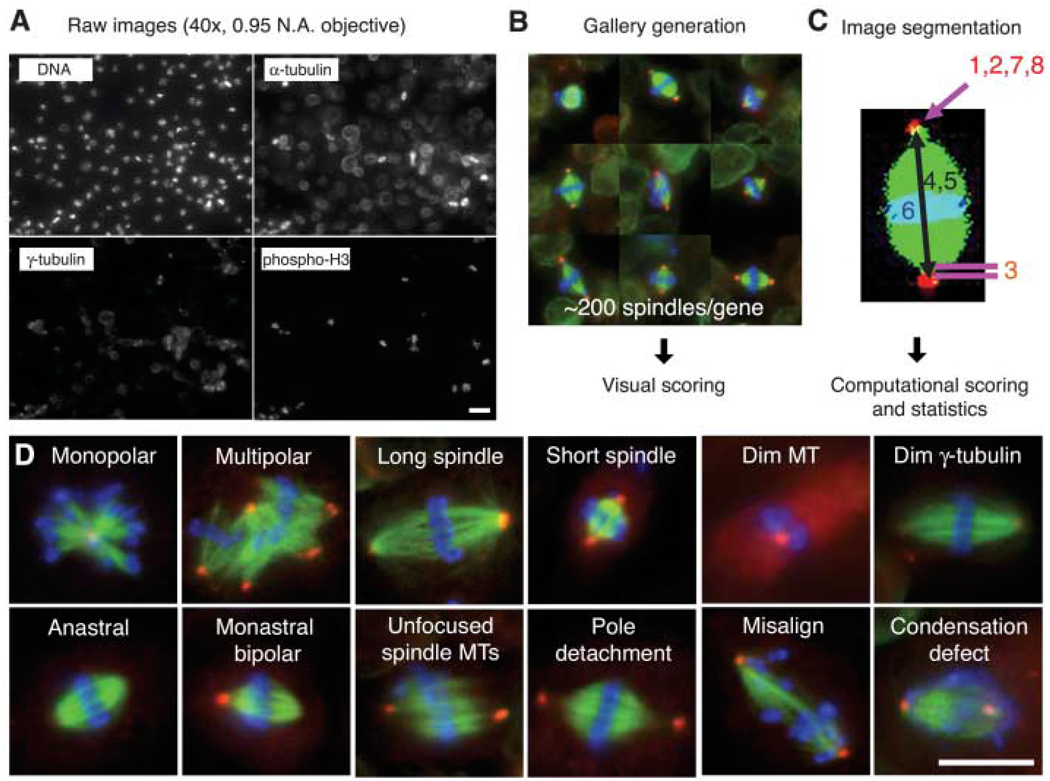
Fig. 1.
An image-based, genome-wide RNAi screening ofmetaphase spindlemorphology. (A) Drosophila S2 cells were treated with dsRNAs of 14,425 genes, along with Cdc27/Apc3 dsRNA, to accumulate metaphase cells. After 4 days, cells were transferred and adhered onto ConA-coated glass-bottom plates. Cells were immunostained for DNA, α-tubulin, γ-tubulin and pH3 and imaged by high-throughput automated microscopy. Scale bar, 20 µm. (B) Mitotic cells were automatically selected by a computer algorithm that detects pH3 staining. The selected ~200 mitotic cells (mostly metaphase) were displayed in a gallery for an observer to score phenotypes. (C) Phenotypes were also analyzed by computer after image segmentation (1, monopolar; 2, multipolar; 3, pole detachment; 4, long spindle; 5, short spindle; 6, misalignment; 7, large γ-tubulin area; 8, dim γ-tubulin) (11). (D) Twelve major phenotypes identified in the screen. Scale bar, 10 µm.