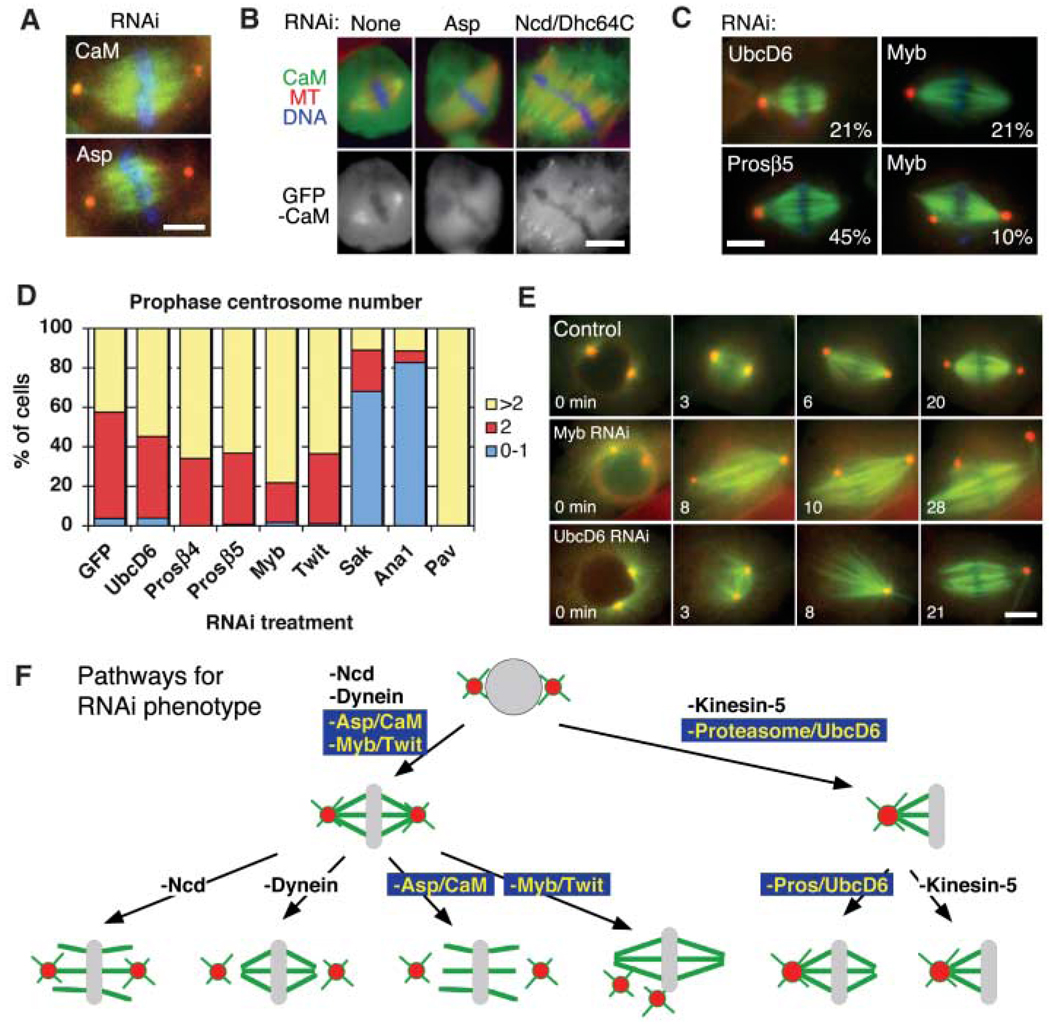
Fig. 3.
Genes required for centrosome separation and association to spindle poles. (A) Identical kinetochore-fiber unfocusing and centrosome-detachment phenotypes were observed after Asp and calmodulin (CaM) RNAi. See movie S2. (B) GFP-CaM was localized at spindle poles in an Asp-dependent manner, but was still detected at unfocused poles induced by Ncd/Dhc64C RNAi. (C) Monastral bipolar spindles produced by UbcD6, proteasome subunit β5, or Myb RNAi (percentage of cells with phenotype indicated; 3% to 8% for untreated cells). Centrosome detachment also was detected after Myb RNAi. (D) At prophase, most proteosome (Prosβ4 and Prosβ5), UbcD6, Myb, and Twit RNAi cells (n > 65) had >1 centrosome; Sak, Ana1 (centriole duplication), and Pav (cytokinesis) served as control RNAis that decrease or increase centrosome numbers. (E) Time-lapse imaging of γ-tubulin-GFP (red) and mCherry-tubulin (green) from prophase to metaphase shows bipolar spindle formation, and then centrosome detachment after Myb RNAi (80%) (n = 20), and immediate centrosome fusion after NEB for UbcD6 RNAi (25%) (n = 20). See movie S3. Scale bars, 5 µm. (F) Models for various spindle-pole phenotypes in response to various protein depletions.