Figure 4.
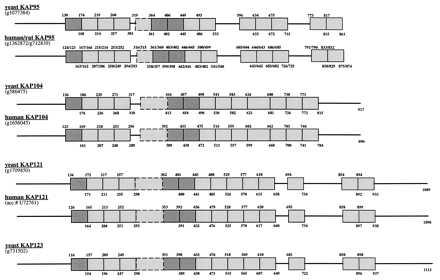
Distribution of HEAT motifs in the β-Kaps. HEAT motifs are indicated as boxes, and non-HEAT-containing sequences are represented by thick lines. The entire human and yeast ORFs were aligned by clustalv (27) in pairs. A window of about 40 amino acids, matching the HEAT consensus (11), then was used to match the ORF pairs with a high gap penalty, to score for individual HEAT motifs. The alignment windows were evaluated individually on the basis of matches to the residues identified previously as being important for the secondary structure. Mismatches of one important residue per α-helix were allowed as long as the secondary structure prediction was unaffected. Secondary structure predictions were used based on the sspred method (29) to confirm the primary sequence alignments. The darkly shaded boxes represent HEAT motifs that are unambiguously homologous in the various β-Kaps (data not shown). The dashed boxes represent HEAT motifs that are not close matches to the universal HEAT consensus (Fig. 3) but are predicted to assume a similar secondary structure (11, 19, 20). Each HEAT repeat is bounded by numbers above and below the box that indicate the beginning and ending amino acid for that particular repeat. Accession numbers/protein identification numbers are indicated below the protein names.
