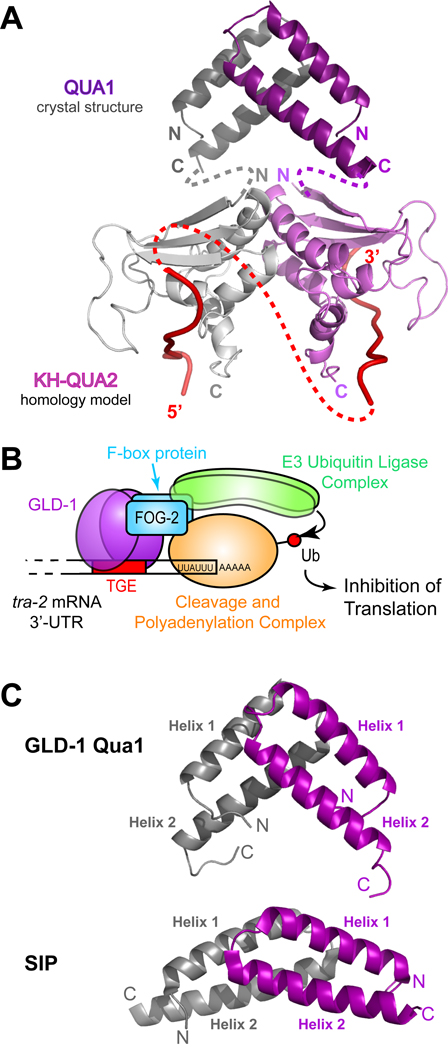
Figure 8. Model of a GLD-1 STAR dimer bound to RNA and model of the GLD-1 translation repression complex.
(A) Model of a GLD-1 STAR dimer. The Qua1 crystal structure is represented as dark gray and purple ribbons. The KH and Qua2 regions are a homology model based on the SF-1 NMR structure (Liu et al., 2001) and are shown as light gray and purple ribbons. The interface between the KH-Qua2 domains was modeled based on the Nova KH3 dimer (Ramos et al., 2002). The connectivity between Qua1 and the KH-Qua2 model is shown schematically as dashed gray and purple lines. Red ribbons represent RNA, and the connectivity of RNA between protomers is depicted as a dashed red line. (B) Model of the GLD-1 translation repression complex on the RNA. GLD-1 recruits an E3 ubiquitin ligase via its interaction with the F-box protein FOG-2. The cleavage and polyadenylation complex may be a potential target for ubiquitination. (C) Comparison of the GLD-1 Qua1 (top) and SIP (bottom) homodimerization domains. The two protomers within the homodimers are colored in gray and purple.