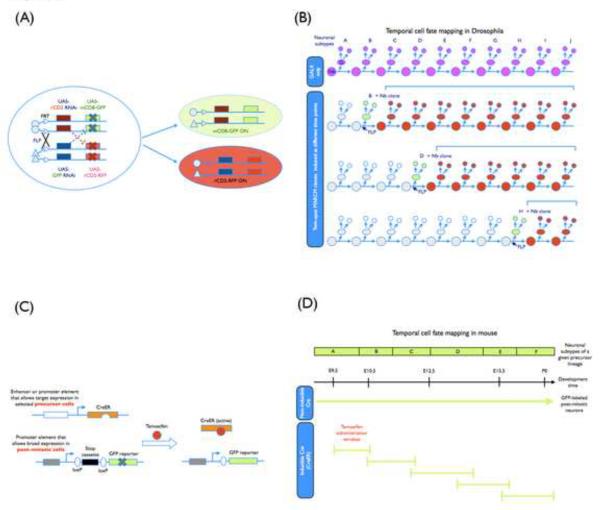
Figure 2.
(A) A schematic representation of genetic elements used in the twin-spot MARCM system. With the adoption of two sets of reporters and corresponding silencers (miRNA-based suppressors against two reporter genes in the current design) that have been placed on opposing homologous chromosome arms and distal to the recombination site, the paired Nb and two-cell clones can be labeled differentially at the same time in a mosaic brain after FRT/FLP-mediated mitotic recombination. Symbol “X” indicates the suppression of reporter gene expression. (B) Temporal cell fate mapping with twin-spot MARCM analysis. For a given neuronal lineage, a selected GAL4 driver was used to label the sequentially produced neural subtypes (A–J subtypes shown in the figure). By including this GAL4 driver in twin-spot MARCM, the temporal identity of individual marked two-cell clones generated at different developmental time points can be unambiguously determined based on the size and composition of their sister Nb clones, allowing high-resolution birth order mapping. (C) A schematic representation of genetic elements used in the inducible genetic fate mapping. Two transgenes are involved. One encodes an inducible site-specific recombinase CreER, which is a fusion protein of Cre recombinase with a tamoxifen-responsive Estrogen Receptor ligand-binding domain. Only after tamoxifen administration, CreER activity is turned on, providing a temporal control over the recombination event. Moreover, CreER expression under the control of a distinct promoter (or enhancer) element is restricted to specific precursors. The other transgenic element encodes an inheritable reporter whose expression depends on Cre-mediated excision of a stop signal and persistently marks the progeny of CreER-positive precursors. (D) Temporal cell fate mapping with the inducible genetic fate mapping in mouse. For a given neuronal precursor lineage, the sequentially derived neural subtypes (A–F types labeled by a non-inducible Cre activity) can be selectively labeled by the CreER activity depending on the time window of tamoxifen administration. Their birth order could be subsequently deduced.