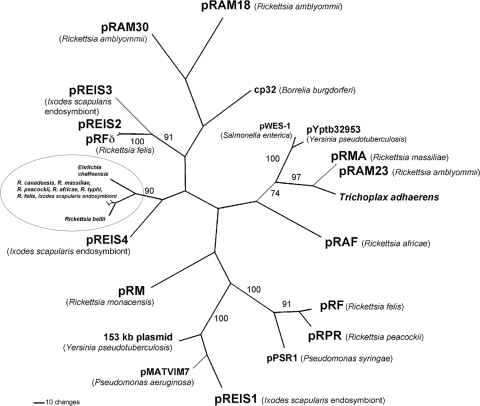
FIG. 6.
Maximum parsimony phylogenetic tree (unrooted) of rickettsial ParA proteins. The scale bar indicates the number of amino acid changes within branch lengths. Bootstrap scores are indicated by numerals. GenBank accession numbers for the sequences used are as follows (in parentheses). Plasmid-encoded ParA sequences were from pRAM18 (GU322808), pRAM23 (GU322807), pRAF (YP_002845772), pREIS1, -2, -3, and -4 (EER20867, EER20809, EER20748, and NZ_GG688316 [translated nucleotides 10818 to 11573]), pRF (YP_247439), pRM (YP_001967398), pRMA (YP_001497196), pRPR (YP_002921997), cp32 (AAL60459), pPSR1 (NP_940697), pWes-1 (YP_002332254), pYptb32953 (YP_068544), pMATVIM7 (YP_001427366), and a 153-kb plasmid (YP_001393406). The Trichoplax adhaerens contig sequence was under accession no. EDV19036. Chromosome-encoded ParA sequences were from E. chaffeensis (ABD44691), R. africae (YP_002844799), R. bellii (YP_538520), R. canadensis (YP_001491792), R. felis (YP_246133), I. scapularis endosymbiont (ZP_04698724), R. massiliae (YP_001498948), R. peacockii (ACR47329), and R. typhi (YP_067042).