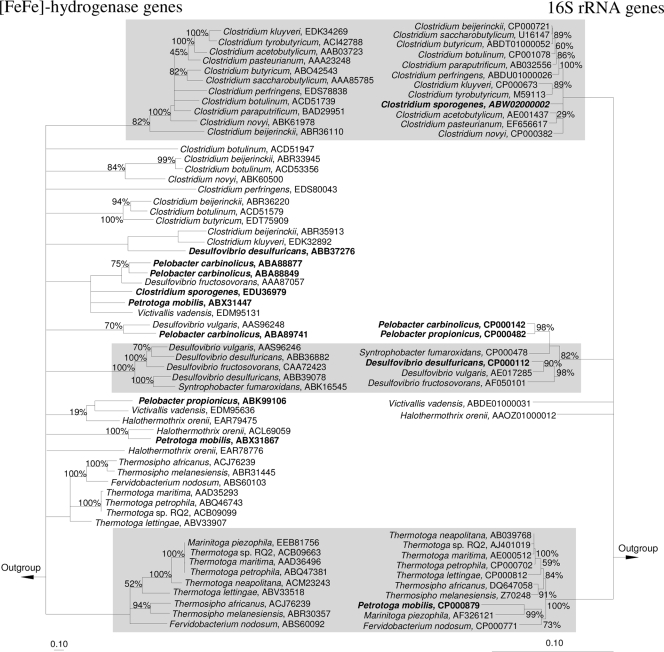
FIG. 3.
Comparison of [Fe-Fe]-hydrogenase- and 16S rRNA gene-based phylogenetic trees (residues 35 to 389 of the D. vulgaris hydrogenase) (see Fig. 1). Both trees are consensus trees calculated and displayed as described for Fig. 2. Consistent monophyletic groups between both trees are shaded in gray. Examples of hydrogenases and hosts with inconsistent phylogenies are in boldface. Bars indicates 0.1 estimated change per amino acid or nucleotide. See Fig. S1 in the supplemental material for trees that include all presently available pure-culture [Fe-Fe]-hydrogenases and corresponding 16S rRNA genes.