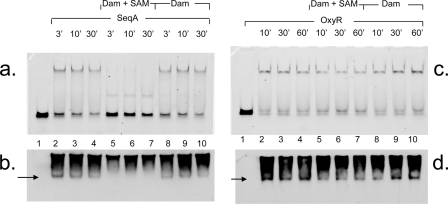
FIG. 1.
Dam methylation rapidly disrupts existing SeqA/HM-DNA complexes, but OxyR/HM-DNA complexes are more stable. (a and c) Results of EMSA after the addition of 7 pmol SeqA (a, lanes 2 to 10) or OxyR (c, lanes 2 to 10) with 0.1 pmol agn43 HM-DNA. Lane 1, DNA only. Complexes were allowed to form for 10 min, after which no protein (lanes 2 to 4), 0.5 pmol Dam supplemented with 64 nM SAM (lanes 5 to 7), or Dam without SAM (lanes 8 to 10) was added. (b and d) After incubation for the times indicated, complexes were analyzed by EMSA gel. The protein identity in the complex was confirmed by Western blot analysis with anti-SeqA (b) or anti-OxyR (d). Arrows in b and d indicate the positions of DNA-protein complexes, and smears represent excess free protein.