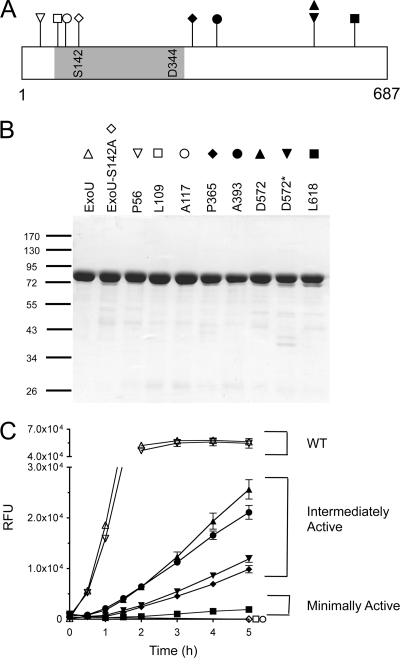
FIG. 3.
Purified ExoU-LINKER proteins and phospholipase activity. (A) Location of ExoU-LINKER molecules chosen for analysis using recombinant proteins. The symbol above each location site in panel A corresponds to the symbol used for each ExoU derivative in panels B and C. The gray region represents the patatin-like phospholipase A2 domain of ExoU. The catalytic sites are marked within the gray region of the diagram. The ExoU-LINKER molecules chosen for analysis include P56 (▿), L109 (□), A117 (○), S142A (⋄), P365 (♦), A393 (•), D572 (▴), D572* (▾), and L618 (▪). Not depicted in the diagram is WT rExoU (▵). Two insertions were obtained at D572, but with different sequences. (B) SDS-PAGE analysis of ExoU-LINKER recombinant proteins used in the fluorescence-based ExoU activity assay. Each well of the Coomassie-stained gel contains approximately 1.5 μg of recombinant protein. The sizes of the molecular weight markers (in thousands) are indicated on the left. (C) Phospholipase activity of recombinant proteins was measured in the fluorescence-based ExoU activity assay. The assay reaction included 0.16 μM ExoU/ExoU-LINKER, 6.3 μM bSOD1, 50 mM MOPS (pH 6.3), 50 mM NaCl, 750 mM MSG (pH 6.3), and 30 μM PED6. RFU were measured every 15 min for 2 h and every h thereafter for a total of 5 h. Recombinant L109 and rA117 possessed the same activity curve as a noncatalytic molecule, S142A. Each point represents the average of 3 independent experiments, with error bars for standard error.