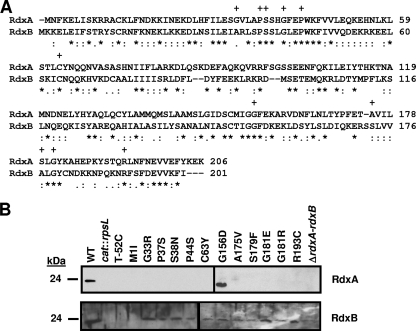
FIG. 2.
Comparison of RdxA and RdxB amino acid sequences and production of RdxA and RdxB in in vitro- and in vivo-isolated Mtzr C. jejuni 81-176 mutants. (A) ClustalV alignment of the amino acid sequences of RdxA and RdxB from C. jejuni 81-176. Asterisks (*) indicate identical residues, semicolons (:) indicate highly conserved residues, and periods (.) indicate weakly conserved residues. Plus signs (+) above residues of RdxA indicate residues changed by point mutations in Mtzr C. jejuni isolates after in vitro or in vivo growth. (B) Immunoblot analysis of RdxA and RdxB production in whole-cell lysates of Mtzr C. jejuni 81-176 isolates with point mutations in rdxA isolated after in vitro or in vivo growth. The wild-type strain is 81-176 Smr (DRH212) (WT), and the cat::rpsL mutants used in the immunoblots are DAR521 (81-176 Smr rdxA::cat-rpsL) and DAR562 (81-176 Smr rdxB::cat-rpsL), respectively. Most Mtzr isolates are identified by the type of change occurring in the protein sequence after spontaneous mutation. T-52C refers to an Mtzr isolate with a nucleotide change occurring 52 bases upstream of the rdxA start codon. ΔrdxA-rdxB refers to an Mtzr isolate with a deletion of DNA encompassing the 3′ end of rdxA through the 5′ end of rdxB.