FIG. 4.
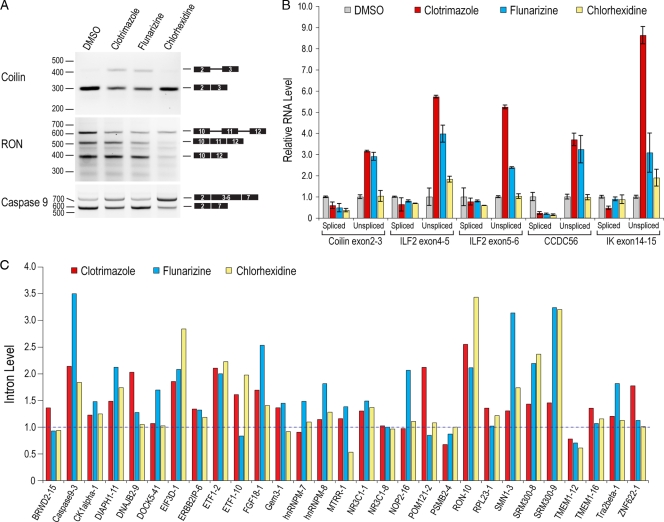
Splicing modulators have distinct effects on constitutive and alternative splicing of endogenous genes in cells. (A) RT-PCR analysis of total RNA extracted from HeLa cells treated with clotrimazole, flunarizine, and chlorhexidine for 6 h. Results are shown for constitutive pre-mRNA splicing of endogenous coilin intron 2 and SR protein-regulated alternative splicing of exon 11 of RON and exons 3 to 6 of caspase 9. (B) Real-time qPCR analysis of the samples shown in panel A. For each endogenous gene, a set of primers was designed to distinguish between exon-exon junction (spliced) and exon-intron junction (unspliced). Coilin intron 2 and ILF2 introns 4 and 5 are major (U2 dependent), whereas CCDC56 intron 1 and IK intron 14 are minor (U12 dependent). Three independent replicates were used for each treatment, and data are presented relative to the level for DMSO, which was set to 1. (C) Real-time qPCR analysis of introns from various endogenous genes, as described for panel A. For each transcript, the intron tested is indicated as a number after the gene name. Each bar represents the average of results from three measurements, and data are presented relative to the level for DMSO, which was set to 1. All qPCR measurements were normalized to the level for β-actin.