Abstract
Recognition of invading viruses by the host is elicited by cellular sensors which trigger signaling cascades that lead to type I interferon (IFN) gene expression. Retinoic acid-inducible gene I (RIG-I) has emerged as a key receptor for the detection of viral RNA in the cytosol, inducing IFN-mediated innate immune responses to limit viral replication through its interaction with MAVS (also called IPS-1, CARDIF, or VISA). Upon the recognition of viral RNA, the Lys-172 residue of RIG-I undergoes ubiquitination induced by tripartite motif protein 25 (TRIM25), an essential protein for antiviral signal transduction. Here we demonstrate that phosphorylation represents another regulatory mechanism for RIG-I-mediated antiviral activity. Using protein purification and mass spectrometry analysis, we identified three phosphorylation sites in the amino-terminal caspase recruitment domains (CARDs) of RIG-I. One of these residues, Thr-170, is located in close proximity to Lys-172, and we speculated that its phosphorylation may affect Lys-172 ubiquitination and functional activation of RIG-I. Indeed, a RIG-I mutant carrying a phosphomimetic Glu residue in place of Thr-170 loses TRIM25 binding, Lys-172 ubiquitination, MAVS binding, and downstream signaling ability. This suggests that phosphorylation of RIG-I at Thr-170 inhibits RIG-I-mediated antiviral signal transduction. Immunoblot analysis with a phospho-specific antibody showed that the phosphorylation of the RIG-I Thr-170 residue is present under normal conditions but rapidly declines upon viral infection. Our results indicate that Thr-170 phosphorylation and TRIM25-mediated Lys-172 ubiquitination of RIG-I functionally antagonize each other. While Thr-170 phosphorylation keeps RIG-I latent, Lys-172 ubiquitination enables RIG-I to form a stable complex with MAVS, thereby inducing IFN signal transduction.
The host's immediate response to viral infections relies on pattern recognition receptors (PRRs) that sense nucleic acids or other conserved structural components of invading viruses. These sensors subsequently initiate signaling cascades leading to the production of type I interferons (IFNs) and other cytokines, which in turn mediate innate immune responses to limit viral replication. The host has evolved at least two classes of PRRs for the detection of viruses, differing fundamentally with respect to their cellular localization: the transmembrane-localized Toll-like receptors (TLRs) and the cytosolic receptors retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) (3, 31). While TLRs detect incoming virions in endosomes or phagosomes of specialized immune cells, such as plasmacytoid dendritic cells (pDCs), RIG-I and MDA5 sense actively replicating viruses in the cytoplasm of most nonimmune cells (4, 28, 31, 32). RIG-I is activated by different types of viral RNA, such as 5′-triphosphate single-stranded RNA and double-stranded RNA (dsRNA), while MDA5 is activated by dsRNA (12, 23, 32). In line with this, the generation of RIG-I or MDA5 knockout mice demonstrated the critical role of RIG-I in IFN production following infection with paramyxoviruses, flaviviruses, and influenza viruses. In contrast, MDA5 was shown to recognize picornaviruses (14, 17). In addition, it was recently shown that RIG-I is involved in the recognition of cytosolic dsDNA of various DNA viruses, including adenovirus, herpes simplex virus 1 (HSV-1), and Epstein-Barr virus (EBV) (1, 7). Specifically, cellular DNA-dependent RNA polymerase III transcribes viral dsDNA into 5′-triphosphate RNA species that activate RIG-I, thereby leading to type I IFN induction.
RIG-I and MDA5 are RNA helicases characterized by a conserved domain structure comprising two N-terminal caspase recruitment domains (CARDs) and a central DExD/H-box ATPase/helicase domain. In addition, RIG-I possesses a C-terminal regulatory/repressor domain (RD) (24, 32). The C-terminal RD of RIG-I, containing a zinc coordination site, binds viral RNA in a 5′-triphosphate-dependent manner (8, 12, 23, 27). RNA binding subsequently leads to the stimulation of the ATPase/helicase. Helicase activity is presumed to induce RIG-I conformational alteration and multimerization, thereby unmasking the N-terminal CARDs. The CARDs of RIG-I and MDA5 then mediate the interaction with the CARD of mitochondrial antiviral signaling protein (MAVS; also known as IPS-1, CARDIF, or VISA) (15, 18, 25, 29). MAVS functions as an adaptor, linking the sensors RIG-I and MDA5 to the kinases TBK1 (TANK-binding kinase 1) and IKK-ɛ (inhibitor of nuclear factor Iκ kinase-ɛ), which phosphorylate interferon-regulatory factors 3 and 7 (IRF3/7) (21). Upon phosphorylation, IRF3/7 dimerizes, translocates into the nucleus, and subsequently induces IFN-α/β gene expression in concerted action with NF-κB and ATF-2/c-Jun transcription factors.
Tight regulation of immune signaling pathways is essential for a successful immune response against viral infections. Whereas positive regulatory mechanisms lead to the rapid activation of IFN signaling upon viral infection, negative regulatory mechanisms are required to prevent unwanted or excessive production of IFNs or proinflammatory cytokines. It has become increasingly evident that posttranslational modifications represent an important cellular mechanism in regulating or fine-tuning RIG-I signal-transducing ability. RIG-I activity is negatively regulated by Lys-48-linked ubiquitination, leading to RIG-I degradation, as well as by ISG15 conjugation (5, 16). Furthermore, we have previously reported that the RIG-I N-terminal CARDs undergo Lys-63-linked ubiquitination induced by tripartite motif 25 (TRIM25) E3 ligase and that TRIM25-mediated ubiquitination of RIG-I at Lys-172 is essential for efficient RIG-I-MAVS interaction, and thereby for the ability of RIG-I to elicit antiviral responses against RNA virus infection (10). In addition, a splice variant of RIG-I that is unable to interact with TRIM25 loses CARD ubiquitination and downstream signaling activity, demonstrating the crucial role of TRIM25-induced ubiquitination for RIG-I signaling function (9).
Here we show that phosphorylation of the RIG-I N-terminal CARDs represents another regulatory mechanism to control its antiviral activity. Using protein purification and mass spectrometry (MS) analysis, we identified three phosphorylation sites in the RIG-I CARDs. Mutational analysis demonstrated that phosphorylation of the RIG-I Thr-170 residue inhibits TRIM25 binding and RIG-I CARD ubiquitination, thereby suppressing the signal-transducing ability of RIG-I. Using a Thr-170 phospho-specific antibody, we show that RIG-I undergoes robust phosphorylation at Thr-170 under normal conditions but that this phosphorylation rapidly decreases upon viral infection. We thus propose that Thr-170 phosphorylation and TRIM25-mediated Lys-172 ubiquitination of RIG-I functionally antagonize each other to tightly regulate RIG-I signal-transducing activity.
MATERIALS AND METHODS
Cell culture and transfection.
RIG-I−/− mouse embryonic fibroblasts (MEF) were described previously (10). HEK293T, MEF, and Vero cells were grown in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% fetal calf serum (FCS) (Gibco-BRL), 2 mM l-glutamine, and 1% penicillin-streptomycin. Transient transfections were performed with calcium phosphate (Clontech) according to the manufacturer's instructions.
Plasmid construction.
All constructs for the transient expression of RIG-I, MAVS, and TRIM25 proteins in mammalian cells were derived from the pEBG GST fusion vector or the pEF-IRES-puro expression vector. DNA fragments corresponding to the coding sequences of the RIG-I, MAVS, and TRIM25 genes were amplified from template DNA by PCR and subcloned into plasmid pEBG, between KpnI and NotI sites, or into pEF-IRES-puro, between AflII and NotI sites. V5- and Flag-tagged RIG-I, MAVS, and TRIM25 constructs were expressed from a modified pEF-IRES-puro vector encoding C-terminal V5 and Flag tags, respectively. All RIG-I mutant constructs used in this study were generated by site-directed mutagenesis, using primers containing the desired mutation, and were completely sequenced using an ABI Prism 377 automatic DNA sequencer to verify the presence of the mutation. Expression plasmids encoding enhanced green fluorescent protein (EGFP) fused to human IRF3 (pEGFP-IRF3) and Flag-tagged human IRF3 (pCMV2-Flag-IRF3) were described previously (6, 19).
Generation of stable 293T and RIG-I−/− MEF cells.
To generate HEK293T cells stably expressing empty vector, wild-type RIG-I (RIG-I WT), or its mutants, cells were transfected with DNA for pEF-IRES-puro vector, pEF-IRES-puro-RIG-I-Flag, or its mutants, followed by selection with 2 μg/ml puromycin (Sigma). For the generation of RIG-I−/− MEF stably complemented with RIG-I WT or its mutants, DNA encoding RIG-I or its mutants was cloned into the pBabe-puro vector. Each plasmid was transfected into EcoPack2-293 cells (Clontech) to produce the retroviruses. For the control, an empty pBabe-puro vector was transfected. The RIG-I−/− MEF cells were transduced with retroviruses carrying each construct and selected with 0.8 μg/ml puromycin (Sigma).
Reagents and antibodies.
For the detection of RIG-I Thr-170 phosphorylation by immunoblot analysis, HEK293T cells were treated with 100 nM phosphatase inhibitor calyculin A (Invitrogen) for 45 min before being harvested. For immunoblotting, the following primary antibodies were used: anti-V5 (1:5,000) (Invitrogen), anti-Flag (M2; 1:5,000) (Sigma), anti-glutathione S-transferase (anti-GST) (1:10,000) (Sigma), anti-ubiquitin (P4D1; 1:500) (Santa Cruz), anti-polyubiquitin (Lys-63-linkage-specific) (1:200) (Biomol), monoclonal anti-TRIM25 (1:5,000) (BD Biosciences), monoclonal anti-RIG-I (Alme-1; 1:1,000) (Alexis), anti-IRF3 (1:1,000) (Santa Cruz), anti-β-actin (Abcam), and anti-phospho-threonine (1:500) (Cell Signaling Technology). The phospho-specific RIG-I Thr-170 antibody was generated by immunizing rabbits with the phospho-peptide Cys-SDKENWPK(p)TLKLALEKER covalently coupled to keyhole limpet hemocyanin (KLH). For purification purposes, a second group of rabbits were immunized with the non-phospho-peptide Cys-SDKENWPKTLKLALEKER. The presence of phospho-specific immunoreactivity was detected by enzyme-linked immunosorbent assay (ELISA), using both phosphorylated and nonphosphorylated peptides. Phospho-peptide-specific antibodies were purified by first passing the antibodies over immobilized nonphosphorylated peptide to remove antibodies that were reactive to nonphosphorylated epitopes. The nonabsorbed fraction was then passed over a column of immobilized phospho-peptide. After extensive washing, the retained immunoglobulins were eluted, dialyzed, and concentrated.
Luciferase reporter assay.
HEK293T cells were seeded into six-well plates. Twenty-four hours later, the cells were transfected with 500 ng of IFN-β luciferase reporter plasmid and 500 ng constitutive β-galactosidase (β-Gal)-expressing pGK-β-gal. In addition, 10 ng of plasmid encoding RIG-I 2CARD-GST fusions or 500 to 2,000 ng of plasmid encoding Flag-RIG-I mutant genes was transfected. Thirty-six to 40 h after transfection, whole-cell lysates were prepared and subjected to a luciferase assay (Promega). Luciferase values were normalized to β-galactosidase to measure transfection efficiency.
Confocal immunofluorescence microscopy.
HEK293T cells stably expressing empty vector, Flag-RIG-I, or its mutants were seeded into Lab-Tek II eight-well chamber slides (Nalge Nunc International). The following day, the cells were transfected with 200 ng per well of plasmid encoding IRF3-EGFP, followed by infection with 80 hemagglutinin (HA) units/ml Sendai virus (SeV) (Cantell strain; Charles River) 24 h later. Twenty hours after infection, cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.2% (vol/vol) Triton X-100 for 15 min, and blocked with 10% goat serum in phosphate-buffered saline (PBS) for 1 h at room temperature. To detect RIG-I protein, a monoclonal Flag antibody (Sigma) was diluted (1:500) in PBS with 1% goat serum and incubated for up to 2 h at room temperature. After incubation, cells were washed extensively with PBS and then incubated with goat anti-mouse Alexa Fluor 568-conjugated secondary antibody (Molecular Probes, Portland, OR) diluted (1:500) in PBS for 1 h at room temperature, followed by extensive washing with PBS. Confocal microscopy was performed using a Leica TCS SP laser scanning microscope (Leica Microsystems) fitted with a 100× Leica objective (PL Apo; 1.4 numerical aperture [NA]) and Leica imaging software. Images were collected at a resolution of 512 by 512 pixels. The stained cells were optically sectioned along the z axis, and the images in the different channels (photomultiplier tubes) were collected simultaneously. The step size on the z axis varied from 0.2 to 0.8 μm to obtain 4 to 8 slices per imaged file. The images were transferred to a Macintosh G4 computer (Apple Computer, CA), and Photoshop (Adobe) was used to render the images.
Native PAGE.
Native PAGE was performed as described previously (9).
GST pull-down assay, immunoprecipitation, and immunoblot analysis.
HEK293T cells were lysed in NP-40 buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% [vol/vol] NP-40, protease inhibitor cocktail [Roche], and Ser/Thr-phosphatase inhibitor cocktail [Sigma]), followed by centrifugation at 13,000 rpm for 20 min. For GST pull-down assays, postcentrifugation supernatants were mixed with a 50% slurry of glutathione-conjugated Sepharose beads (Amersham Biosciences), and the binding reaction mix was incubated for 3 to 4 h at 4°C. For immunoprecipitation, supernatants were incubated with the indicated antibodies at 4°C for 4 to 12 h. Generally, 1 to 2 μg of antibody was added to 1 ml of cell lysate. After addition of protein A/G agarose beads, the incubation was continued for 2 h. Immunoprecipitates were extensively washed with lysis buffer and eluted with SDS loading buffer by boiling for 5 min. For immunoblotting, whole-cell lysates or precipitated proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). The membranes were incubated in blocking buffer (PBS, 5% [wt/vol] nonfat dry milk powder) for 1 h at room temperature, followed by incubation with primary antibody diluted in blocking buffer for 1 h at room temperature or at 4°C overnight. Membranes were washed extensively with PBST (PBS, 0.1% Tween 20), followed by incubation with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. The proteins were visualized by use of an enhanced chemiluminescence reagent (Pierce) and were detected with a phosphorimager (Fuji LAS-4000).
In vivo GST pull-down and MS analysis.
Forty-eight hours after transfection with GST-RIG-I 2CARD, HEK293T cells were collected and lysed with NP-40 buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% [vol/vol] NP-40) supplemented with Complete protease inhibitor cocktail (Roche) and Ser/Thr-phosphatase inhibitor cocktail (Sigma). Postcentrifugation supernatants were mixed with a 50% slurry of glutathione-conjugated Sepharose beads (Amersham Biosciences), and the binding reaction mix was incubated for 4 h at 4°C. Precipitates were washed extensively with lysis buffer. Proteins bound to glutathione beads were eluted and separated in a NuPAGE 4 to 12% Bis-Tris gradient gel (Invitrogen). After Coomassie staining, protein bands corresponding to the ubiquitinated and nonubiquitinated forms of GST-RIG-I 2CARD were excised and separately analyzed by ion-trap mass spectrometry at the Harvard Taplin Biological Mass Spectrometry Facility, Boston, MA.
Excised gel bands were cut into approximately 1-mm3 pieces. The samples were reduced with 1 mM dithiothreitol for 30 min at 60°C and then alkylated with 5 mM iodoacetamide for 15 min in the dark at room temperature. Gel pieces were then subjected to a modified in-gel trypsin digestion procedure (26). Gel pieces were washed and dehydrated with acetonitrile for 10 min, followed by removal of acetonitrile. Pieces were then completely dried in a Speed-Vac apparatus. Rehydration of the gel pieces was performed with 50 mM ammonium bicarbonate solution containing 12.5 ng/μl modified sequencing-grade trypsin (Promega, Madison, WI) at 4°C. Samples were then placed in a 37°C room overnight. Peptides were extracted by removing the ammonium bicarbonate solution, followed by one wash with a solution containing 50% acetonitrile and 5% acetic acid. The extracts were then dried in a Speed-Vac apparatus (∼1 h). The samples were stored at 4°C until analysis. On the day of analysis, the samples were reconstituted in 5 μl of high-performance liquid chromatography (HPLC) solvent A (2.5% acetonitrile, 0.1% formic acid). A nano-scale reverse-phase HPLC capillary column was created by packing 5-μm C18 spherical silica beads into a fused silica capillary (100-μm inner diameter × 12-cm length) with a flame-drawn tip (22). After equilibration of the column, each sample was pressure loaded off-line onto the column. The column was then reattached to the HPLC system. A gradient was formed, and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). As each peptide was eluted, it was subjected to electrospray ionization and then placed in an LTQ-Orbitrap mass spectrometer (ThermoFinnigan, San Jose, CA). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identities) were determined by matching sequences from protein or translated-nucleotide databases with the acquired fragmentation patterns by use of Sequest software (ThermoFinnigan, San Jose, CA). The modification of 79.9663 mass units of serine, threonine, and tyrosine was included in the database searches to determine phosphopeptides. Each phosphopeptide that was identified by the Sequest program was also manually inspected to ensure confidence.
IFN-β ELISA and VSV replication.
HEK293T and complemented RIG-I−/− MEF cells were seeded into a 12-well plate and mock infected or infected with either SeV (Cantell strain; Charles River) (50 HA units/ml) or ΔNS1 A/PR8/34 (H1N1) influenza virus (multiplicity of infection [MOI] of 2). Twenty-four hours after infection, supernatants were collected and analyzed for IFN-β production by ELISA (PBL Biomedical Laboratories). For viral replication assays, stable HEK293T or RIG-I−/− MEF cells were seeded into six-well plates and infected with vesicular stomatitis virus expressing EGFP (VSV-EGFP) at an MOI of 0.5. Twenty-six to 42 h after infection, the culture medium was harvested and the virus yield determined by plaque assay on Vero cells.
Homology modeling.
Automated protein structure homology modeling prediction servers Phyre and Swiss-Model were used to generate a model for the RIG-I second CARD-like domain, with similar solutions. The MAVS CARD-like domain (PDB accession no. 2vgq) was used as a template for the alignment mode prediction in Swiss-Model. The ubiquitin structure was obtained from the Protein Data Bank (PDB accession no. 1UBQ). Pymol (Delano Scientific) was used to generate the final visualization for the structural model.
RESULTS
Phosphorylation of RIG-I CARDs.
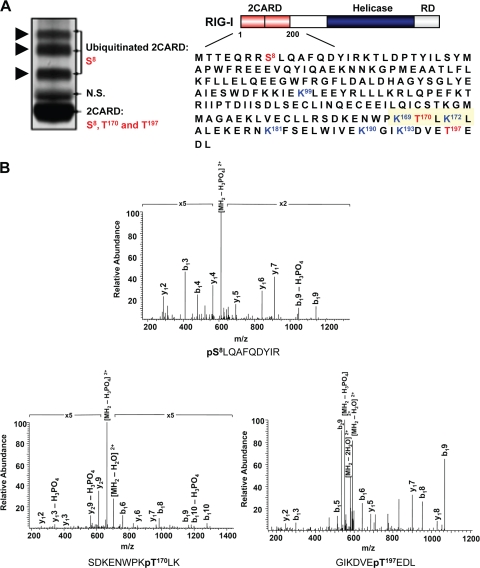
A recent series of studies demonstrated that ubiquitination plays a pivotal role in controlling RIG-I antiviral activity (5, 9-11, 20). As part of our continuous effort to define the molecular mechanisms that regulate RIG-I signal-transducing ability, we sought to identify additional posttranslational modifications of the RIG-I N-terminal CARDs. To this end, we transiently expressed a mammalian GST fusion construct of the RIG-I CARDs (GST-RIG-I 2CARD) in HEK293T cells. As previously shown (10), Coomassie staining of purified GST-RIG-I 2CARD resulted in four detectable species: nonubiquitinated GST-RIG-I 2CARD, with a size of 44 kDa, and three slower-migrating polypeptides, with apparent molecular masses of 52, 60, and 68 kDa, representing the ubiquitinated forms of RIG-I CARDs. The in vivo ubiquitinated and nonubiquitinated species of GST-RIG-I 2CARD were then analyzed separately by multidimensional liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). This showed that the RIG-I CARDs carry phosphorylations at Ser-8, Thr-170, and Thr-197 (Fig. 1A and B). Notably, whereas the ubiquitinated and nonubiquitinated species of RIG-I CARDs carry a phosphorylations at Ser-8, the Thr-170 and Thr-197 phosphorylations were detected only in the nonubiquitinated RIG-I 2CARD (Fig. 1A, left panel). No tyrosine phosphorylation sites of the RIG-I CARDs were detected under the same conditions (data not shown). These results demonstrate that the N-terminal CARDs of RIG-I undergo in vivo phosphorylation at Ser-8, Thr-170, and Thr-197.
FIG. 1.
Phosphorylation of RIG-I CARDs. (A) RIG-I 2CARD phosphorylation sites determined by MS. (Left) Coomassie-stained purified GST-RIG-I 2CARD (amino acids 1 to 200). (Right) RIG-I structure and amino acid sequence of the N-terminal CARDs. Arrows indicate the ubiquitinated forms of GST-RIG-I 2CARD. Phosphorylation sites identified by MS are highlighted in red. The blue lysine residues indicate the sites of ubiquitination (10). The area around Thr-170 is highlighted in yellow. N.S., nonspecific band. (B) MS/MS spectra of tryptic phosphopeptides pSLQAFQDYIR, SDKENWPKpTLK, and GIKDVEpTEDL, which identified phosphorylation sites at Ser-8, Thr-170, and Thr-197. p, phosphorylation. b- and y-ion designations are shown (2).
Detection of RIG-I Thr-170 phosphorylation by a phospho-specific p-Thr-170 antibody.
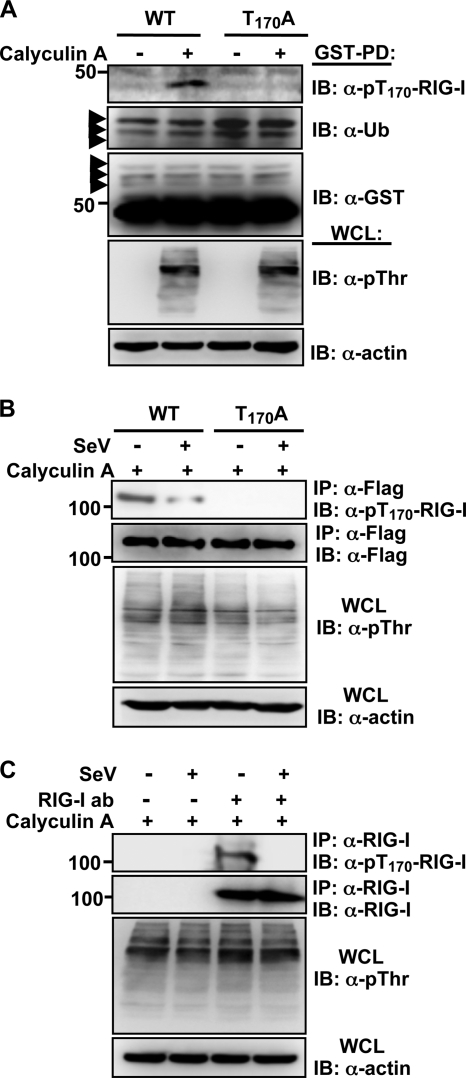
It is intriguing that the Thr-170 residue of RIG-I is located in close proximity to the Lys-172 residue, the major site for TRIM25-mediated ubiquitination (Fig. 1A, right panel). To demonstrate the in vivo Thr-170 phosphorylation of RIG-I, an 18-amino-acid peptide [S162DKENWPK(p)TLKLALEKER179] containing the phosphorylated Thr-170 residue was used to generate a phosphorylation state-specific p-Thr-170 antibody. This antibody was shown to be highly specific for the phosphorylated Thr-170 epitope by ELISA (data not shown). Furthermore, the p-Thr-170 antibody efficiently reacted with GST-RIG-I 2CARD WT and Flag-RIG-I WT but not with GST-RIG-I 2CARD T170A and Flag-RIG-I T170A mutants (Fig. 2A and B). Remarkably, Thr-170 phosphorylation was readily detected in exogenously and endogenously expressed RIG-I under normal conditions, but in striking contrast, it markedly declined upon SeV infection (Fig. 2B and C). These results suggest that RIG-I undergoes robust phosphorylation at the Thr-170 residue and that this phosphorylation markedly decreases upon viral infection.
FIG. 2.
Detection of Thr-170 phosphorylation of RIG-I in vivo by using a phospho-Thr170 RIG-I antibody. (A) Thr-170 phosphorylation of RIG-I 2CARD. Forty-eight hours after transfection with GST-RIG-I 2CARD WT or the T170A mutant, HEK293T cells were mock treated or treated with calyculin A. Whole-cell lysates (WCLs) were subjected to GST pull-down assay (GST-PD), followed by immunoblotting (IB) with anti-pT170-RIG-I, anti-ubiquitin (anti-Ub), or anti-GST antibody. WCLs were further used for immunoblotting with anti-phosphothreonine (anti-pThr) and anti-actin antibodies. Arrows indicate the ubiquitinated bands. (B) RIG-I phosphorylation at Thr-170 decreases upon viral infection. After transfection with Flag-tagged RIG-I WT or the T170A mutant, HEK293T cells were mock infected or infected with SeV (50 HA units/ml) for 4 h. WCLs were subjected to immunoprecipitation (IP) with an anti-Flag antibody, followed by immunoblotting with anti-pT170-RIG-I and anti-Flag antibodies. WCLs were also used for immunoblotting with anti-p-Thr and anti-actin antibodies. (C) Thr-170 phosphorylation of endogenous RIG-I. HEK293T cells were mock infected or infected with SeV (50 HA units/ml) for 6 h. WCLs were subjected to immunoprecipitation with an anti-RIG-I antibody, followed by immunoblotting with anti-pT170-RIG-I or anti-RIG-I antibody. WCLs were also used for immunoblotting with anti-pThr or anti-actin antibody.
T170E mutation suppresses RIG-I ubiquitination, MAVS binding, and signaling activity.
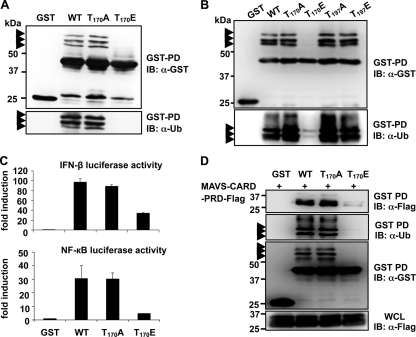
The fact that the Thr-170 phosphorylation of RIG-I is exclusively detected in the nonubiquitinated form of RIG-I 2CARD indicates that this phosphorylation might play a critical role in regulating RIG-I ubiquitination and, thereby signal-transducing ability. To test this hypothesis, the Thr-170 residue was replaced with alanine (T170A) to mimic nonphosphorylation or with glutamic acid (T170E) to mimic constitutive phosphorylation. In addition, we also generated RIG-I 2CARD mutants in which Thr-197 was replaced with alanine (T197A) or glutamic acid (T197E). GST-RIG-I 2CARD mutants were then tested for their ubiquitination levels and signal-transducing abilities. As shown in Fig. 3A and B, GST-RIG-I 2CARD T170E exhibited a near-complete loss of ubiquitination compared with GST-RIG-I 2CARD WT, whereas GST-RIG-I 2CARD T170A showed ubiquitination equivalent to that seen with GST-RIG-I 2CARD WT. In contrast, the T197A and T197E mutations had little to no effect on the ubiquitination level of RIG-I 2CARD (Fig. 3B). In correlation with its lack of ubiquitination, the GST-RIG-I 2CARD T170E mutant exhibited a markedly reduced ability to initiate IFN-β- and NF-κB-mediated promoter activation compared with that of GST-RIG-I 2CARD WT and the T170A mutant (Fig. 3C). The CARDs of RIG-I have been shown to efficiently bind to the MAVS CARD to elicit downstream signal transduction (15, 18, 25, 29). GST pull-down analysis showed that GST-RIG-I 2CARD WT and the T170A mutant efficiently interacted with the Flag-tagged CARD-proline-rich domain of MAVS (Flag-MAVS-CARD-PRD). In striking contrast, the MAVS-binding ability of GST-RIG-I 2CARD T170E was reduced ∼86% compared with that of GST-RIG-I 2CARD WT (Fig. 3D). These results collectively indicate that the T170E mutation, which mimics phosphorylation of the Thr-170 residue, strongly suppresses the ubiquitination, MAVS interaction, and signal-transducing ability of RIG-I.
FIG. 3.
Phosphorylation of RIG-I at Thr-170 inhibits RIG-I ubiquitination, MAVS binding, and signaling activity. (A and B) T170E mutation abolishes RIG-I CARD ubiquitination. Forty-eight hours after transfection with GST, GST-RIG-I 2CARD WT, or the indicated mutants, HEK293T WCLs were used for GST pull-down assays (GST-PD), followed by immunoblotting with an anti-GST or anti-Ub antibody. Arrows indicate the ubiquitinated bands. (C) T170E mutation decreases RIG-I downstream signaling. GST-RIG-I fusion constructs, together with IFN-β or NF-κB luciferase and constitutive β-Gal-expressing pGK-β-gal, were expressed in HEK293T cells. Luciferase and β-galactosidase values were determined as described previously (9). Data represent the means ± standard deviations (SD) (n = 3). (D) T170E mutation strongly decreases RIG-I binding to MAVS. HEK293T cells were transfected with MAVS-CARD-PRD-Flag, together with GST or GST-RIG-I 2CARD fusion constructs. WCLs were subjected to GST pull-down assays (GST-PD), followed by immunoblotting with an anti-Flag, anti-Ub, or anti-GST antibody. MAVS-CARD-PRD expression was determined by immunoblotting with an anti-Flag antibody. Arrows indicate the ubiquitinated bands.
T170E mutation abolishes RIG-I antiviral activity.
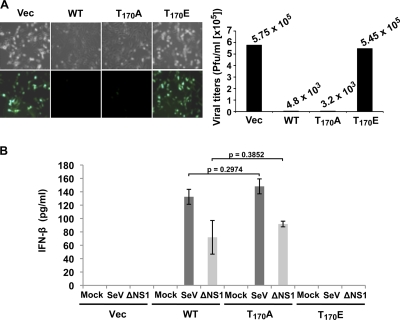
To determine the role of Thr-170 phosphorylation in the regulation of RIG-I activity against RNA virus infection, we tested the replication capacity of VSV-EGFP in RIG-I−/− MEF stably expressing vector, RIG-I WT, RIG-I T170E, or RIG-I T170A (Fig. 4A). RIG-I WT and T170A mutant expression drastically suppressed VSV-EGFP replication; VSV-EGFP titers were ∼120- and 180-fold lower in RIG-I−/− MEF expressing RIG-I WT and the T170A mutant, respectively, than in vector-containing RIG-I−/− MEF (Fig. 4A). In contrast, RIG-I−/− MEF expressing the RIG-I T170E mutant had similar viral titers to those in RIG-I−/− MEF complemented with vector (Fig. 4A). In line with these results, RIG-I−/− MEF stably expressing RIG-I WT or the RIG-I T170A mutant highly induced IFN-β production upon infection with SeV or ΔNS1 A/PR8/34 influenza virus, while RIG-I−/− MEF expressing empty vector or RIG-I T170E showed almost no IFN-β production under the same conditions (Fig. 4B). In summary, these results indicate that the RIG-I T170E mutant completely loses the ability to induce antiviral IFN production.
FIG. 4.
T170E mutation abolishes RIG-I antiviral activity. (A) VSV-EGFP replication in complemented RIG-I−/− MEF. RIG-I WT, RIG-I T170A, or RIG-I T170E was stably expressed in RIG-I−/− MEF, and these cells were infected with VSV-EGFP at an MOI of 0.5. Twenty-six hours after infection, virus replication and titers were determined by GFP expression (left) and plaque assay (right), respectively. (B) IFN-β production in complemented RIG-I−/− MEF. RIG-I−/− MEF stably expressing RIG-I WT or the indicated RIG-I mutants were infected with 50 HA units/ml SeV or with ΔNS1 A/PR8/34 influenza virus at an MOI of 2. Twenty-four hours after infection, IFN-β production in the supernatants was determined by ELISA. Statistical analysis was performed by unpaired Student's t test.
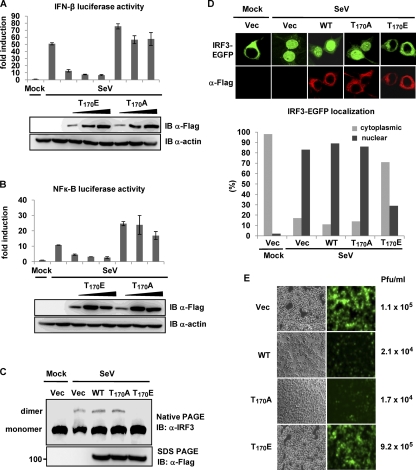
The RIG-I T170E mutant acts as a dominant inhibitor of RIG-I-mediated antiviral signaling.
We next examined the effects of the RIG-I T170E and T170A mutants on RIG-I signal transduction activity. HEK293T cells were transiently transfected with IFN-β or NF-κB luciferase constructs together with increasing amounts of Flag-RIG-I T170E and T170A mutants, followed by SeV infection (Fig. 5A and B). RIG-I T170E expression potently suppressed SeV-induced IFN-β or NF-κB promoter activation in a dose-dependent manner. In contrast, IFN-β and NF-κB promoter activities were slightly increased by exogenously expressed RIG-I T170A (Fig. 5A and B). To further delineate the inhibitory effect of the T170E mutant on the RIG-I-mediated downstream signaling cascade, we examined virus-induced dimerization and nuclear translocation of IRF3. HEK293T cells were cotransfected with Flag-IRF3 and RIG-I WT, RIG-I T170A, or RIG-I T170E, followed by SeV infection (Fig. 5C). This showed that while IRF3 was primarily present as a monomer in mock-infected cells, SeV infection led to the efficient dimerization of IRF3 in cells transfected with empty vector, RIG-I WT, or RIG-I T170A. In contrast, ectopic expression of the RIG-I T170E mutant markedly suppressed the SeV-induced dimerization of IRF3 (Fig. 5C). Furthermore, while IRF3-EGFP was localized almost exclusively in the cytoplasm of mock-infected cells, IRF3-EGFP nuclear translocation was readily detected upon SeV infection in cells expressing vector, RIG-I WT, or RIG-I T170A (Fig. 5D). In contrast, expression of the RIG-I T170E mutant strongly decreased the virus-induced nuclear translocation of IRF3-EGFP: IRF3-EGFP was localized in the cytoplasm in 71% of the cells expressing RIG-I T170E upon viral infection (Fig. 5D, lower panel). Finally, we examined the effects of the RIG-I T170A and T170E mutants on VSV-EGFP replication (Fig. 5E). RIG-I T170E expression detectably increased VSV-EGFP replication compared with vector expression. In contrast, the replication of VSV-EGFP was markedly decreased in cells expressing RIG-I WT or the RIG-I T170A mutant (Fig. 5E). These results collectively indicate that the RIG-I T170E mutant acts as a dominant inhibitor of RIG-I-mediated antiviral signaling.
FIG. 5.
RIG-I T170E mutant inhibits RIG-I-mediated antiviral signaling. (A and B) RIG-I T170E inhibits SeV-induced IFN-β or NF-κB promoter activation. HEK293T cells were transfected with vector or increasing amounts of Flag-tagged RIG-I T170E or RIG-I T170A, together with IFN-β (A) or NF-κB (B) luciferase and pGK-β-gal. Twenty-four hours after transfection, cells were mock treated or infected with SeV (50 HA units/ml), and luciferase activity was determined as described in the legend to Fig. 3C. Data represent the means ± SD (n = 3). WCLs were used for immunoblotting with anti-Flag and anti-actin antibodies. (C) RIG-I T170E inhibits virus-induced IRF3 dimerization. Twenty-four hours after transfection with Flag-IRF3, Flag-RIG-I WT, Flag-RIG-I T170A, or Flag-RIG-I T170E, HEK293T cells were mock treated or infected with SeV (80 HA units/ml) for 20 h. WCLs were either used for native PAGE followed by immunoblotting with an anti-IRF3 antibody or subjected to SDS-PAGE followed by immunoblotting with anti-Flag antibody. (D) RIG-I T170E inhibits virus-induced nuclear translocation of IRF3. Twenty-four hours after transfection with IRF3-EGFP, HEK293T cells stably expressing vector, Flag-RIG-I, Flag-RIG-I T170A, or Flag-RIG-I T170E were mock treated or infected with SeV (80 HA units/ml) and stained with anti-Flag antibody (red). More than 200 cells in which both RIG-I WT or a mutant and IRF3-EGFP were expressed were counted, and the percentage of the cells with nuclear or cytoplasmic IRF3 is shown (lower panel). (E) RIG-I T170E expression increases VSV-EGFP replication. HEK293T cells stably expressing vector (Vec), RIG-I, RIG-I T170A, or RIG-I T170E were infected with VSV-EGFP at an MOI of 0.5. Forty-two hours after infection, virus titers and replication were determined by plaque assay and GFP expression, respectively.
Antagonistic functions of Thr-170 phosphorylation and TRIM25-mediated ubiquitination of RIG-I.
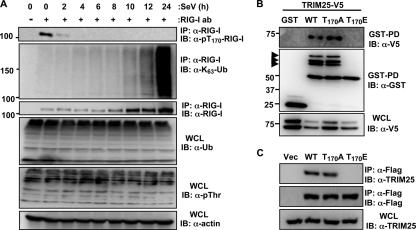
To decipher the molecular mechanism explaining how phosphorylation of the RIG-I Thr-170 residue suppresses TRIM25-mediated ubiquitination, we examined the levels of Lys-63-linked ubiquitination and Thr-170 phosphorylation of endogenous RIG-I at different time points after SeV infection (Fig. 6A). As previously reported (10), the levels of RIG-I ubiquitination were very weak prior to SeV infection but markedly increased upon viral infection, in a time-dependent manner (Fig. 6A). In striking contrast, RIG-I phosphorylation at Thr-170 was readily detected in mock-infected cells but rapidly decreased upon viral infection, such that RIG-I Thr-170 phosphorylation was nearly undetectable at 4 h postinfection or at later time points (Fig. 6A).
FIG. 6.
Antagonistic functions of Thr-170 phosphorylation and TRIM25-mediated ubiquitination of RIG-I. (A) Thr-170 phosphorylation and Lys-63-linked ubiquitination of endogenous RIG-I. HEK293T cells were mock infected or infected with SeV (50 HA units/ml) for the indicated hours. WCLs were subjected to immunoprecipitation with an anti-RIG-I antibody, followed by immunoblotting with anti-pThr-170-RIG-I, anti-Lys-63-polyubiquitin, and anti-RIG-I antibodies. WCLs were immunoblotted with anti-Ub, anti-pThr, or anti-actin antibody. (B and C) T170E mutation abolishes RIG-I-TRIM25 interaction. (B) Forty-eight hours after transfection with GST, GST-RIG-I 2CARD WT, GST-RIG-I 2CARD T170A, or GST-RIG-I 2CARD T170E, together with TRIM25-V5, WCLs were used for GST pull-down assay (GST-PD), followed by immunoblotting with an anti-V5 or anti-GST antibody. WCLs were further immunoblotted with anti-V5. Arrows indicate the ubiquitinated bands. (C) WCLs of HEK293T cells transfected with Flag-tagged RIG-I WT, RIG-I WT T170A, or RIG-I WT T170E were subjected to immunoprecipiation with anti-Flag, followed by immunoblotting with anti-TRIM25 and anti-Flag antibodies. Endogenous TRIM25 expression was determined by immunoblotting with an anti-TRIM25 antibody.
We then tested RIG-I WT, RIG-I T170A, and RIG-I T170E for their ability to interact with TRIM25 (Fig. 6B and C). Consistent with their high ubiquitination levels, GST-RIG-I 2CARD WT and the T170A mutant efficiently interacted with V5-TRIM25 (Fig. 6B). In contrast, the GST-RIG-I 2CARD T170E mutant, showing a near-complete loss of ubiquitination, did not bind V5-TRIM25 under the same conditions (Fig. 6B). In line with this, full-length RIG-I WT and the T170A mutant, but not the RIG-I T170E mutant, strongly bound to endogenous TRIM25 (Fig. 6C). These results strongly suggest that RIG-I phosphorylation at the Thr-170 residue inhibits TRIM25 interaction, thereby suppressing the TRIM25-mediated ubiquitination of RIG-I.
DISCUSSION
Cross talk between phosphorylation and ubiquitination is an emerging theme in the regulation of eukaryotic proteins (13). Our study reveals that the cytosolic viral RNA receptor RIG-I undergoes Ser/Thr phosphorylation at its N-terminal CARDs and that phosphorylation at Thr-170 regulates the Lys-63-linked ubiquitination of RIG-I, thereby regulating its signal-transducing ability. Cross-regulation between phosphorylation and ubiquitination can be either positive or negative, meaning that one modification promotes or inhibits the other. Interestingly, mass spectrometry analysis of the RIG-I CARDs showed that RIG-I modifications by Thr-170 phosphorylation and ubiquitination are mutually exclusive: while ubiquitinated RIG-I CARDs did not show any detectable phosphorylation at Thr-170, nonubiquitinated RIG-I 2CARD robustly underwent Thr-170 phosphorylation (Fig. 1). In addition, replacement of Thr-170 with the phosphomimetic residue Glu almost completely abolished RIG-I CARD ubiquitination, RIG-I MAVS binding, and RIG-I downstream signaling (Fig. 3). Thus, our data indicate that the Thr-170 phosphorylation and Lys-63-linked ubiquitination of RIG-I functionally antagonize each other. These results strongly suggest that while ubiquitination enables RIG-I to form a stable complex with its MAVS downstream partner, thereby inducing antiviral signal transduction (9, 10), phosphorylation of the RIG-I Thr-170 residue keeps RIG-I in an inactive state by suppressing its ubiquitination-dependent activation.
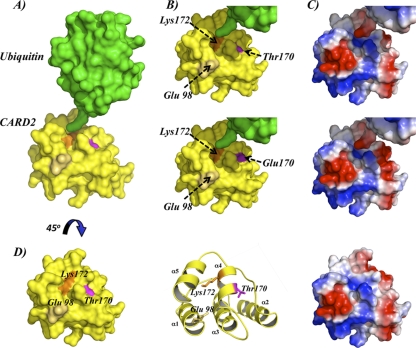
Phosphorylation can negatively regulate ubiquitination in at least two ways, by inhibiting E3 ligase substrate binding or by interfering with ubiquitin attachment. As shown in Fig. 7, homology modeling of the RIG-I second CARD shows that the Thr-170 residue is located in close proximity to the Lys-172 residue, the critical site for TRIM25-mediated ubiquitination and activation of RIG-I (10). It is therefore possible that phosphorylation of the Thr-170 residue sterically affects TRIM25-mediated ubiquitin attachment to the Lys-172 residue by altering the surface charge of this area (Fig. 7B and C). In fact, the introduction of a Glu residue in place of the Thr-170 residue markedly enhances the negative surface charge of this area (Fig. 7B and C, lower panels), thus possibly providing negatively charged repulsion. Interestingly, the fact that the RIG-I T170E mutant did not bind TRIM25 (Fig. 6B and C) suggests that RIG-I Thr-170 phosphorylation may interfere with its TRIM25 interaction. Although our previous work demonstrated that the RIG-I first CARD is necessary and sufficient for TRIM25 binding (9), it is possible that residues located in the RIG-I second CARD allosterically modulate TRIM25 binding. Modeling prediction shows that the Thr-170 residue is located close to the negatively charged Glu-98 residue in the C-terminal part of the short linker region between the RIG-I first and second CARD (Fig. 7B and D). Thus, the negative charge provided by Thr-170 phosphorylation may induce a repulsive effect in the linker region, thereby structurally altering the first CARD, ultimately blocking TRIM25 interaction. It is equally conceivable that the phosphorylated Thr-170 residue forms an ionic interaction with positively charged residues located within the RIG-I first CARD, leading to a structural conformation of the RIG-I tandem CARD in which the first CARD is no longer accessible for interaction with TRIM25. On the other hand, Lys-172 ubiquitination may prevent access of a cellular kinase to phosphorylate Thr-170, which would explain why both posttranslational modifications are mutually exclusive. Further structural analyses of the RIG-I CARDs will be required to determine how RIG-I phosphorylation at Thr-170 affects the tandem CARD structure and TRIM25-mediated ubiquitination of RIG-I.
FIG. 7.
Structural model of RIG-I second CARD-like domain. (A) Structural model of RIG-I second CARD-like domain and ubiquitin in close proximity to Lys-172. Lys-172 is depicted in orange, Thr-170 in pink, and Glu-98 in beige. (B) Surface models of WT RIG-I second CARD (top) and T170E mutant (bottom) in close proximity to the LRLRGG C-terminal motif of ubiquitin. (C) Surface electrostatic potentials of residues shown in panel B. Red and blue depict negative and positive potentials, respectively. (D) Close-up surface view, ribbon representation, and surface electrostatic potentials of RIG-I second CARD-like domain model, with Thr-170 in pink, Lys-172 in orange, and Glu-98 in beige. Glu-98, the first amino acid of the RIG-I second CARD-like domain in close proximity to Lys-172 and Thr-170.
Multisite posttranslational modification, generally referred to as the “protein modification code,” is a key mechanism in controlling the activities of critical molecules in transcription and signal transduction, such as histone proteins or the tumor suppressor p53. In addition to phosphorylation and TRIM25-mediated Lys-63-linked ubiquitination, RIG-I undergoes Lys-48-linked ubiquitination, leading to its degradation as well as conjugation by ISG15, ultimately inhibiting RIG-I's signal-transducing ability (5, 16). Furthermore, it was recently reported that Riplet/REUL is another ubiquitin E3 ligase for Lys-63-linked ubiquitination of RIG-I, thereby promoting RIG-I antiviral signaling (11, 20). Thus, it would be interesting to address how the interplay among these modifications might function to form a code-like multisite modification program for the dynamic control of RIG-I-mediated signal transduction. Future proteomic surveys by mass spectrometry will be directed at decrypting this dynamic “molecular barcode” of RIG-I combinatorial posttranslational modifications.
Several recent studies have suggested a multistep model for RIG-I activation (30). In uninfected cells, the C-terminal RD leads to RIG-I autoinhibition by interacting with both CARD and helicase domains (24). Upon viral infection, 5′-triphosphate RNA binds to the C-terminal RD to induce RIG-I multimerization and activation of its helicase/ATPase (8, 27). ATPase activity is then believed to induce a conformational change in RIG-I, thereby exposing the N-terminal CARDs to enable interaction with downstream partners. Our study indicates that prior to viral infection, the RIG-I second CARD undergoes robust phosphorylation at Thr-170, leading to the inhibition of RIG-I signaling activity. When the GST-RIG-I T170E construct comprising only the tandem CARD was used, it did not undergo ubiquitination and exhibited a strongly reduced downstream signaling activity (Fig. 3A and C). This suggests that the negative charge provided by Thr-170 phosphorylation may impose conformational changes within the tandem CARD, thereby preventing TRIM25 interaction and TRIM25-induced ubiquitination. However, it is also possible that in resting cells the Thr-170 phosphorylation may help to maintain the closed, inactive conformation of RIG-I by stabilizing the interaction between the RD and the CARDs. Thus, as seen with the RIG-I RD, which functions to autoinhibit RIG-I multimerization and CARD signaling, T170 phosphorylation may be an additional way to prevent the CARD-dependent downstream signaling of RIG-I. Further studies are directed toward addressing the molecular details of the role of Thr-170 phosphorylation in RIG-I signaling, including how it affects the structural basis of the CARD itself and the CARD-RD interaction, how it blocks TRIM25 binding, and whether it influences the RNA binding, ATPase activity, or multimerization of RIG-I.
Our study further showed that treatment of cells with the Ser/Thr phosphatase inhibitor calyculin A markedly increased the T170 phosphorylation of RIG-I, indicating that RIG-I T170 phosphorylation is regulated by the dynamic balance between kinase-dependent phosphorylation and phosphatase-dependent dephosphorylation. Furthermore, our results suggest that the kinase(s) responsible for T170 phosphorylation has easy access to the Thr-170 residue in resting cells, whereas phosphatase accessibility to the RIG-I CARDs is significantly limited under the same conditions. Consequently, the negative charge provided by Thr-170 phosphorylation may impose conformational changes within the tandem CARD, thereby preventing TRIM25 interaction and TRIM25-induced ubiquitination. Upon viral infection, however, additional conformational changes within the RIG-I CARDs may then allow access of a phosphatase to remove the T170 phosphorylation. Alternatively, expression of a phosphatase specific for RIG-I T170 may be induced by IFNs. In either scenario, dephosphorylation of the p-T170 residue ultimately permits TRIM25 binding and TRIM25-mediated ubiquitination, allowing RIG-I to form a stable complex with its downstream partner MAVS to trigger IFN-mediated antiviral innate immunity. It will be interesting to investigate how the dynamic balance between RIG-I phosphorylation and dephosphorylation is coordinated to regulate its antiviral signaling activity. Furthermore, the identification of the kinase(s)/phosphatase(s) that regulates phosphorylation of RIG-I Thr-170 will shed new light on how viral infection may modulate the activities of these cellular enzymes in promoting activation of RIG-I and of the IFN-mediated antiviral response.
Acknowledgments
We greatly thank Ross Tomaino (Taplin Mass Spectrometry Facility, Boston, MA) for mass spectrometry analysis and Sean Whelan (Harvard Medical School, Boston, MA) for kindly providing recombinant VSV.
This work was partly supported by U.S. Public Health Service grant RR00168 (M.U.G.), grants CA082057, U19AI083025, and AI083355, the Fletcher Jones Foundation, and the Hastings Foundation (J.U.J. and M.U.G.), and grants U19AI083025 and R01AI46954 and NIAID contract HHSN266200700010C (A.G.-S.).
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Ablasser, A., F. Bauernfeind, G. Hartmann, E. Latz, K. A. Fitzgerald, and V. Hornung. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10:1065-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebersold, R., and D. R. Goodlett. 2001. Mass spectrometry in proteomics. Chem. Rev. 101:269-295. [DOI] [PubMed] [Google Scholar]
- 3.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 4.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arimoto, K., H. Takahashi, T. Hishiki, H. Konishi, T. Fujita, and K. Shimotohno. 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. USA 104:7500-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu, Y. H., J. B. Macmillan, and Z. J. Chen. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui, S., K. Eisenacher, A. Kirchhofer, K. Brzozka, A. Lammens, K. Lammens, T. Fujita, K. K. Conzelmann, A. Krug, and K. P. Hopfner. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169-179. [DOI] [PubMed] [Google Scholar]
- 9.Gack, M. U., A. Kirchhofer, Y. C. Shin, K. S. Inn, C. Liang, S. Cui, S. Myong, T. Ha, K. P. Hopfner, and J. U. Jung. 2008. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. USA 105:16743-16748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gack, M. U., Y. C. Shin, C. H. Joo, T. Urano, C. Liang, L. Sun, O. Takeuchi, S. Akira, Z. Chen, S. Inoue, and J. U. Jung. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916-920. [DOI] [PubMed] [Google Scholar]
- 11.Gao, D., Y. K. Yang, R. P. Wang, X. Zhou, F. C. Diao, M. D. Li, Z. H. Zhai, Z. F. Jiang, and D. Y. Chen. 2009. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One 4:e5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 13.Hunter, T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28:730-738. [DOI] [PubMed] [Google Scholar]
- 14.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 15.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 16.Kim, M. J., S. Y. Hwang, T. Imaizumi, and J. Y. Yoo. 2008. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 82:1474-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loo, Y. M., J. Fornek, N. Crochet, G. Bajwa, O. Perwitasari, L. Martinez-Sobrido, S. Akira, M. A. Gill, A. Garcia-Sastre, M. G. Katze, and M. Gale, Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 19.Ning, S., L. E. Huye, and J. S. Pagano. 2005. Regulation of the transcriptional activity of the IRF7 promoter by a pathway independent of interferon signaling. J. Biol. Chem. 280:12262-12270. [DOI] [PubMed] [Google Scholar]
- 20.Oshiumi, H., M. Matsumoto, S. Hatakeyama, and T. Seya. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J. Biol. Chem. 284:807-817. [DOI] [PubMed] [Google Scholar]
- 21.Paz, S., Q. Sun, P. Nakhaei, R. Romieu-Mourez, D. Goubau, I. Julkunen, R. Lin, and J. Hiscott. 2006. Induction of IRF-3 and IRF-7 phosphorylation following activation of the RIG-I pathway. Cell. Mol. Biol. (Noisy-le-Grand) 52:17-28. [PubMed] [Google Scholar]
- 22.Peng, J., and S. P. Gygi. 2001. Proteomics: the move to mixtures. J. Mass Spectrom. 36:1083-1091. [DOI] [PubMed] [Google Scholar]
- 23.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 24.Saito, T., R. Hirai, Y. M. Loo, D. Owen, C. L. Johnson, S. C. Sinha, S. Akira, T. Fujita, and M. Gale, Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 104:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 26.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 27.Takahasi, K., M. Yoneyama, T. Nishihori, R. Hirai, H. Kumeta, R. Narita, M. Gale, Jr., F. Inagaki, and T. Fujita. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29:428-440. [DOI] [PubMed] [Google Scholar]
- 28.Uematsu, S., and S. Akira. 2007. Toll-like receptors and type I interferons. J. Biol. Chem. 282:15319-15323. [DOI] [PubMed] [Google Scholar]
- 29.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 30.Yoneyama, M., and T. Fujita. 2007. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 282:15315-15318. [DOI] [PubMed] [Google Scholar]
- 31.Yoneyama, M., and T. Fujita. 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 18:545-551. [DOI] [PubMed] [Google Scholar]
- 32.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]