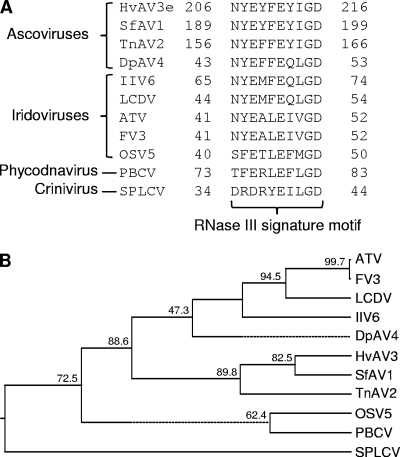
FIG. 1.
Different viruses encode class I RNase III-like proteins. (A) Only RNase III signature motifs in amino acid alignments that are necessary for dsRNA cleavage are shown. The positions of these motifs with respect to amino acid coordinates are variable in different viruses. (B) The full-length amino acid sequences of RNase III from viruses used in panel A were aligned by the CLUSTAL W method based on which a phylogenetic tree was constructed (DNASTAR; MegAlign). A dotted line on the phenogram indicates a negative branch length, a common result of averaging. HvAV-3e (Heliothis virescens AV), SfAV-1 (Spodoptera frugiperda AV), TnAV-2 (Trichoplusia ni AV), DpAV-4 (Diadromus pulchellus AV), IIV-6 (Invertebrate iridescent virus 6), LCDV (Lymphocystis disease virus 1), ATV (Ambystoma tigrinum virus), FV3 (Frog virus 3), OSV 5 (Ostreococcus virus 5), PBCV (Paramecium bursaria chlorella virus), and an RNase III sequence from the RNA virus SPCSV are included in the phenogram. Bootstrap values are shown on the tree.