Abstract
Prion diseases result from the accumulation of a misfolded isoform (PrPSc) of the normal host prion protein (PrPC). PrPSc propagates by templating its conformation onto resident PrPC to generate new PrPSc. Although the nature of the PrPSc-PrPC complex is unresolved, certain segments or specific residues are thought to feature critically in its formation. The polymorphic residue 129 is one such site under considerable study. We combined transmission studies with a novel live cell yeast-based fluorescence resonance energy transfer (FRET) system that models the molecular association of PrP in a PrPSc-like state, as a way to explore the role of residue 129 in this process. We show that a reduction in efficiency of prion transmission between donor PrPSc and recipient PrPC that are mismatched at residue 129 correlates with a reduction in FRET between PrP-129M and PrP-129V in our yeast model. We further show that this effect depends on the different secondary structure propensities of Met and Val, rather than the specific amino acids. Finally, introduction of the disease-associated P101L mutation (mouse- equivalent) abolished FRET with wild-type mouse PrP, whereas mutant PrP-P101L displayed high FRET with homologous PrP-P101L, as long as residue 129 matched. These studies provide the first evidence for a physical alteration in the molecular association of PrP molecules differing in one or more residues, and they further predict that the different secondary structure propensities of Met and Val define the impaired association observed between PrPSc and PrPC mismatched at residue 129.
Keywords: Diseased/Prions, Prions, Protein, Protein/Chemistry, Protein/Conformation, Protein/Protein-Protein Interactions
Introduction
The prion diseases are transmissible neurodegenerative disorders that result from the accumulation of a misfolded isoform (PrPSc) of the normally folded cellular prion protein (PrPC). The former is distinguished from the latter by its insolubility in non-ionic detergents and relative resistance to proteinase K (PK)2 digestion (1). Once generated, PrPSc propagates by templating its conformation onto PrPC, leading to the accumulation of PrPSc and associated central nervous system pathologic features of neuronal death, gliosis, and vacuolation (2). The PrPSc-PrPC interface has not been resolved, although a specific orientation is predicted, based on several lines of evidence suggesting that sequence homology between PrPSc and PrPC at key sites within the molecule are necessary for effective propagation of PrPSc, a feature that forms the basis for the well recognized species barrier to prion transmission (3).
The central region of PrP has been shown to feature prominently in defining the species barrier (4), and within the first β-strand in this central region lies residue 129, a polymorphic site that plays a key role in disease risk and phenotype determination (5, 6). Compared with the general population, homozygosity for either Met or Val is significantly more prevalent in Creutzfeldt-Jakob disease (CJD) (5), whereas the course of disease in 129MV patients is generally more protracted (7, 8). Furthermore, all primary cases of bovine spongiform encephalopathy-related variant (v) CJD are 129MM (9) and mice that express human (Hu) PrP-129M (i.e. Tg(HuPrP-129M)Prnpo/o) were found to be more receptive to vCJD and more faithfully reproduced the phenotype of vCJD than did Tg(HuPrP-129V)Prnpo/o mice (10, 11).
Thus, a key element that determines the efficiency of prion propagation appears to be whether residue 129 of PrPSc and PrPC are the same or different. However, whether this results from an inefficient or altered physical association of PrPSc with PrPC is not known, nor is it known whether the specific amino acids, Met and Val, or their physical properties, determine the nature of the PrPSc-PrPC association. To address these questions, we compared the results from transmission of sCJD to Tg(HuPrP-129M)Prnpo/o and Tg(HuPrP-129V)Prnpo/o mice, with those from co-expression of homologous and heterologous PrP molecules using a novel live cell yeast-based FRET system to model the molecular association of PrP in a PrPSc-like state (12). We show that efficiency of sCJD transmission is enhanced when residue 129 of PrPSc and PrPC match and this correlates with a more ordered molecular association of PrPSc with PrPC, as assessed by FRET. We further find that the effect of 129 occurs in the presence and absence of at least one autosomal dominant mutation (P102L) and this property is determined by their predicted secondary structure propensities. The remarkable consistency of findings between our FRET system and in vivo transmission data support the utility of this system as a novel strategy with which to study the molecular determinants of prion propagation.
MATERIALS AND METHODS
Collection and PRNP Sequencing of sCJD Cases
Tissue samples were obtained from patients followed at the University of Chicago Memory Center, using an IRB approved protocol. DNA was extracted from whole blood samples or frozen brain sections obtained at autopsy, as previously described (13). The entire coding segment of the PRNP gene was sequenced using Big Dye terminator chemistry, as previously described (14). Based on unpublished data,3 only samples carrying PrPSc-Type 1 were used.
Inoculations
Fresh frozen frontal cortex from autopsied sCJD cases were used to prepare a 1% (w/v) brain homogenate in phosphate-buffered saline, as previously described (13), of which 30 μl was intracerebrally inoculated to each mouse. Transgenic (Tg) mice expressing human PrPC with either Met (Tg440) or Val (Tg152) at residue 129 on a mouse PrP knock-out background (i.e. Tg(HuPrP-129M or V)Prnpo/o) were previously constructed and described elsewhere (4). Mice were anesthetized with a xylazine/ketamine mixture, the head fixed in a small animal stereotaxic instrument (Kopf) fitted with mouse ear bars, the scalp was swabbed with alcohol, and a 25-gauge needle attached to a 1-cc syringe filled with inoculum was lowered 3 mm deep directly through the scalp into the parietal lobe. Disease was defined by the presence of at least two signs, including reduced spontaneous movement, scruffy coat, hunched back, or unsteady gait, at which time it was killed for analysis. Well over 20 Tg(HuPrP-129M)Prnpo/o and Tg(HuPrP-129V)Prnpo/o control mice have been inoculated with either 30 μl of phosphate-buffered saline or normal brain homogenates and have not developed prion features or histopathological evidence of prion disease after >600 days.3
Histology
Brains were fixed in 4% formalin for 48 h, then immersed in 97% formic acid for 90 min, washed several times in 4% formalin, dehydrated in xylene and graded alcohol, and saturated with paraffin. The fixed brains were stained with hematoxylin and eosin for assessment of spongiform degeneration.
Generation of PrP Constructs
The mouse PrP coding sequence, extending from residue 23 to 230, which excludes the N-terminal signal sequences for endoplasmic reticulum targeting and the C-terminal glycosylphosphatidylinositol anchor, was previously cloned into the pSP72 (Promega) cloning vector for construction of wild-type (WT) PrP23–230, using standard molecular biological techniques. To limit degradation by yeast cytoplasmic proteases, a serine was added to the N and C termini of the PrP construct. This construct was then subcloned into the p425 and p426 Gal-inducible yeast expression vectors (Promega), prior to mutation of mouse codon 128 using forward and reverse primers flanking codon 128 and the QuikChange II Site-directed Mutagenesis kit (Strategene). For FRET construct generation, the 128 mutants were constructed similarly from PrP23–230::CFP and PrP23–230::YFP templates, as previously described (12). All constructs were confirmed by sequencing. The mutations and primer pairs used to generate them are as follows: 128Val(ATG → GTG), 5′-CTT GGT GGC TAC GTG CTG GGG AGC (+), 5′-GCT CCC CAG CAC GTA GCC ACC AAG (−); 128Ile (ATG → ATT), 5′-CTT GGT GGC TAC ATT CTG GGG AGC (+), 5′-GCT CCC CAG AAT GTA GCC ACC AAG (−); 128Tyr (ATG → TAC), 5′-CTT GGT GGC TAC TAC CTG GGG AGC (+), 5′-GCT CCC CAG GTA GTA GCC ACC AAG (−); 128Leu (ATG → TTG), 5′-CTT GGT GGC TAC TTG CTG GGG AGC (+), 5′-GCT CCC CAG CAA GTA GCC ACC AAG (−); 128Ala (ATG → GCG), 5′-CTT GGT GGC TAC GCG CTG GGG AGC (+), 5′-GCT CCC CAG CGC GTA GCC ACC AAG (−); 128Lys (ATG → AAG), 5′-CTT GGT GGC TAC AAG CTG GGG AGC (+), 5′-GCT CCC CAG CTT GTA GCC ACC AAG (−); P101L (CCC → CTC), 5′-CCC CCC AAT CAG TGG AAC AAG CTC AGC AAA CCA AAA (+), 5′-TTT TGG TTT GCT GAG CTT GTT CCA CTG ATT GGG GGG (−).
PrP Expression in Yeast
The yeast strain W303 (MATa ade2-can1-100 his3-12,16 leu2-3,112, trp1-1 ura 3-1) with a pep4− genotype, with decreased endogenous protease activity, was used. The p425 and p426 Gal vectors, selectable with medium lacking Leu and Ura, respectively, contain a regulatable element induced by galactose and restricted by glucose. For transformation, yeast colonies were picked from plates, grown overnight at 30 °C to mid-log phase, washed, then incubated with the construct of interest plus polyethylene glycol, LiAc, and carrier DNA for 30 min at 30 °C, followed by a 25-min heat shock at 42 °C, and then spread onto selection plates prepared from glucose dropout media.
Solubility Assay
Yeast expressing the construct of interest were grown to mid-late log phase (A600 ∼0.7), centrifuged at 3,000 × g for 5 min, washed in 10 mm EDTA, and digested with 1 mg/ml of zymolyase 100T for at least 2 h at 30 °C. Spheroplasts were pelleted and lysed in 200 μl of 10% Sarkosyl in TEN buffer (40 mm Tris-Cl, pH 7.5, 1 mm EDTA, pH 8.0, 150 mm NaCl) with protease inhibitors for 5 min on ice. Insoluble PrP was pelleted by centrifugation at 16,000 × g for 30 min at 4 °C. The supernatant was removed and the pellet washed with 50 μl of 10% Sarkosyl, centrifuged again, and resuspended in 200 μl of 1% sulfobetaine in phosphate-buffered saline. Equal fractions were subjected to SDS-PAGE.
FRET
Fluorescence microscopy and photobleaching FRET experiments were performed with an Olympus IX81 microscope using appropriate Chroma filters. PrP23–230::CFP donor was co-expressed with a PrP23–230::YFP acceptor and yeast were scanned for the presence of both cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) signals. CFP or CFP fusion proteins were detected by 436 nm excitation/480 nm emission, and YFP or YFP fusion proteins were detected by 500-nm excitation/535-nm emission filters. Photographs of yeast containing aggregates were taken at 5-s intervals with 100-ms exposure times under constant illumination for 2 min. Image series were saved in stacks and the illumination intensity profile of the aggregates were analyzed across the time (Z) series using ImageJ (NIH) software. The decay of the donor fluorescence signal was fit with a single exponential decay curve and Tau was generated using Igor Pro (Wavemetrics) software using the equation Y0 = Aexp(−invτ*X). The presence of both YFP and CFP signals for a given sample was confirmed before analysis. Colocalized proteins were expressed in greater than 80% of aggregates. To control for differences in lamp intensity across data collection sittings, which can alter the absolute bleach rate of samples, only two donor/acceptor pairs were measured at a time.
Statistical Analyses
All statistical analyses were performed using GraphPad 4 (Prism) software. All reported p values are based on one-way analysis of variance of the group data, followed by Dunn's Multiple Comparison post hoc test, unless otherwise stated.
Protease Digestion Assay
For yeast lysates, PK digestions were performed on samples from the 1% sulfobetaine resuspensions of the 16,000 × g pellet fractions of PrP. Approximately 400 ng of total protein was digested with 10–60 μg/ml of PK at 37 °C for 1 h. A mixture of protease inhibitors (chymostatin (60 μg/ml), pepstatin A (0.7 μg/ml), and aprotinin (2 μg/ml)) were included in the reactions to protect against yeast proteases. For human and mouse brain, a 10% brain homogenate was prepared in lysis buffer (20 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, 0.5% sodium deoxycholate) from frozen brain cortex, without protease inhibitors, and digested with 20 μg/ml of PK at 37 °C for 1 h. All reactions were terminated with 2 mm phenylmethylsulfonyl fluoride.
Western Blots
PK-resistant PrP from brain homogenates was separated by SDS-PAGE on a 14% gel, as previously described (12). Membranes were probed with 3F4 antibody (gift of Richard Kascsak, Staten Island, NY) at a 1:3000 dilution. For PrP from yeast, a fraction of the yeast pellet resuspended in 1% sulfobetaine was separated on a 16% gel and probed with R1 antibody (InPro, South San Francisco, CA) at a 1:5000 dilution. Horseradish peroxidase-conjugated anti-human or anti-mouse secondary antibody (Pierce) was used to detect R1 and 3F4, respectively. Blots were treated with West Pico ECL (Thermo Scientific, Rockford, IL) and captured with a Bio-Rad Alpha Document Imager.
RESULTS
Rate of Transmission of sCJD Depends on Residue 129
We first compared the efficiency of transmission of human (Hu) 129M prions with 129V prions to Tg(HuPrP-129M)Prnpo/o and Tg(HuPrP-129V)Prnpo/o mice, hereafter referred to as Tg(129M) and Tg(129V), respectively. Four cases each of sCJD homozygous for either Met (i.e. 129MM) or Val (129VV) were prepared as 1% brain homogenates in phosphate-buffered saline and intracerebrally inoculated to an equal number (between 5 and 9) of Tg(129M) and Tg(129V) mice. The latency to disease onset, or incubation period, a surrogate measure of transmission efficiency, was determined. Disease was clinically defined by the appearance of at least two symptoms of prion disease in mice, as defined under “Materials and Methods,” and was confirmed by the histological presence of spongiform degeneration, and the biochemical detection of PK-resistant PrP.
All mice inoculated with sCJD(129MM) developed disease, although the incubation period in Tg(129M) mice was significantly shorter than that in Tg(129V) mice that received the same inoculum (Table 1). For example, case 1 produced clinical disease in Tg(129V) mice after ∼294 days, compared with ∼201 days in Tg(129M) mice. Similarly, case 4 transmitted to Tg(129V) mice in ∼230 days and Tg(129M) mice in ∼162 days. Thus, although mice expressing human PrPC carrying either Met or Val at residue 129 were receptive to PrPSc-129M, a mismatch between PrPSc and PrPC at position 129 resulted in a significant delay in the onset of disease (Table 1). These results compare with those of others (10, 15).
TABLE 1.
Prion transmission to Tg(HuPrP)Prnpo/o mice
| Case No. | Codon 129 | Tg(HuPrP-129V) |
Tg(HuPrP-129M) |
||
|---|---|---|---|---|---|
| ni/nsa | IPb | ni/ns | IP | ||
| days | days | ||||
| 1 | MM | 6/6 | 294 ± 11 | 6/6 | 201 ± 16 |
| 2 | MM | 5/5 | 198 ± 2 | 5/5 | 130 ± 12 |
| 3 | MM | 5/5 | 240 ± 1 | 5/5 | 160 ± 6 |
| 4 | MM | 5/5 | 230 ± 15 | 6/6 | 162 ± 4 |
| 5 | VV | 5/5 | 226 ± 5 | 5/6 | 434 ± 7 |
| 6 | VV | 7/7 | 266 ± 2 | 7/8 | 396 ± 17 |
| 7 | VV | 7/7 | 190 ± 4 | 8/9 | 392 ± 21 |
| 8 | VV | 6/6 | 232 ± 6 | 5/7 | 417 ± 12 |
a ns, number sick; ni, number of mice inoculated.
b IP, incubation period.
To ensure that Tg(129M) mice are not inherently more susceptible to sCJD prions than Tg(129V) mice, we compared the incubation periods of Tg(129M) and Tg(129V) mice inoculated with sCJD(129VV). In this case, Tg(129V) mice were significantly more receptive to sCJD(129VV) compared with Tg(129M) mice. On average, the incubation periods were 48% shorter in Tg(129V) mice. For example, case 7 produced disease by ∼190 days in Tg(129V) mice, compared with ∼362 days in Tg(129M) mice (Table 1).
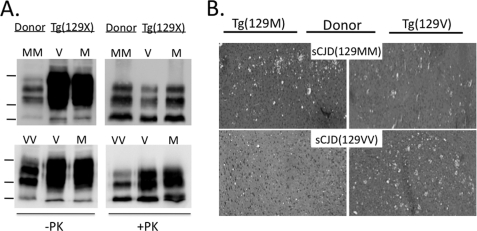
To confirm prion disease, brain homogenates were prepared from more than 80% of mice from each group, and digested with 20 μg/ml of PK, submitted to 14% SDS-PAGE and immunoblotted with the α- HuPrP monoclonal antibody 3F4 (16), to detect PK-resistant PrP (rPrPSc) (Fig. 1A). All symptomatic mice displayed rPrPSc. Interestingly, although mice were killed at a similar stage of disease and the samples were adjusted to the same total protein concentrations, the level of rPrPSc was generally higher in mice that carried the same residue 129 as the human inoculum, supporting a more efficient conversion with homologous PrPC. The pattern of rPrPSc recovered from mice displayed the same molecular phenotype as the human source inoculum. We used only sCJD carrying PrPSc-Type 1, defined primarily by a migration rate of 21 kDa of the unglycosylated fraction of rPrPSc, and this molecular phenotype was propagated in all affected Tg(129M) and Tg(129V).
FIGURE 1.
Transmission of sCJD to Tg(129V) and Tg(129M) mice. A, Western blots of rPrPSc from brain homogenates of the original human sCJD(129MM) and sCJD(129VV) donors and a representative Tg(129V) or Tg(129M) recipient mouse. Samples were freshly prepared from frozen frontal cortex as 10% (w/v) brain homogenate in lysis buffer. Total protein was normalized and subjected to 20 μg/ml of PK for 30 min at 37 °C, probed with monoclonal antibody 3F4. Markers on the left represent 37, 25, and 20 kDa, top to bottom. All mice were clinically sick, as defined under “Materials and Methods.” B, spongiform degeneration evident in hematoxylin and eosin (H&E) stainings of sections of the frontal cortex taken from sick Tg(129M) (left sections) and Tg(129V) (right sections) mice inoculated with brain homogenate from sCJD(129MM) (top sections) or sCJD(129VV) (bottom sections), as indicated. PrP plaques were not present in any brain region.
Histological examination of the brains of all mice confirmed the presence of spongiform degeneration. In general, the degree of spongiform degeneration was greater when the PrPC of recipient mice and the HuPrPSc inoculum carried matching residues at position 129 (Fig. 1B).
PrP-129M and PrP-129V Produce PrPSc-like Protein in Yeast
The results of the transmission studies led us to question whether a mismatch between PrPSc and PrPC at residue 129 directly impacts the nature of their association. To address this, we employed a previously described heterologous yeast expression system that supports the generation of PrPSc-like protein (12, 17). When the N-terminal signal sequence for endoplasmic reticulum entry and the C-terminal glycosylphosphatidylinositol anchor signal sequence are deleted, full-length PrP (i.e. PrP23–230) expression is diverted to the yeast cytosol, where it acquires the physical characteristics of PrPSc, including detergent insolubility and relative PK resistance. Because PrP expressed in the yeast cytosol produces aggregates that acquire PK resistance, newly synthesized PrPC is predicted to associate with the preformed aggregate and is converted to the PrPSc-like isoform. Thus, this model approximates the association of PrPC with PrPSc.
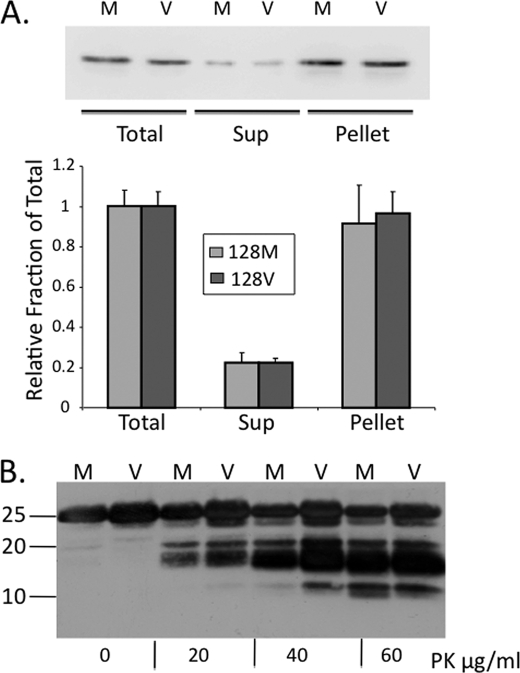
Although we previously showed that PrP-129M acquires properties of PrPSc (12) it was necessary to determine whether PrP-129V also supports its production. Saccharomyces cerevesiae were transformed to express mouse PrP23–230 with either Met or Val at position 128 (the murine equivalent of residue 129 in HuPrP). After 16 h of growth, the yeast were detergent lysed, PrP was pelleted by centrifugation at 16,000 × g, and the insoluble fraction was subjected to a range of PK from 0 to 60 μg/ml. PrP23–230 bypasses the post-translational processing in the endoplasmic reticulum and Golgi, resulting in a single unglycosylated ∼25-kDa fraction. The major fraction of PrP recovered from yeast expressing either PrP-128M or PrP-128V, was insoluble (Fig. 2A), and each PrP displayed PK resistance (Fig. 2B), supporting both to generate PrPSc-like protein in this system.
FIGURE 2.
PrP insolubility and PK resistance of PrP-128M and PrP-128V in the yeast cytosol. A, yeast cells expressing mouse sequence PrP-128M and PrP-128V were lysed in TEN buffer containing 10% Sarkosyl, from which equal aliquots of the total, supernatant (Sup), and pellet fractions, following 16,000 × g centrifugation, were subjected to Western analysis. PrP was probed with the C-terminal anti-mouse PrP F(ab) R1 antibody (residues 225–231). Solubility profile of each PrP was determined as the relative density of the supernatant signal relative to the total signal from each of 3 assays, using a Bio-Rad Alpha Document Imager and Quantity One® (Bio-Rad) software. The pellet represents nearly 90% of the fraction, suggesting rapid conversion of newly synthesized soluble PrP to the insoluble aggregate. B, Western blot of PrP-128M (M) and PrP-128V (V) expressed in the yeast cytosol and treated with PK (0 to 60 μg/ml) for 1 h at 37 °C. Prominent PK-resistant fragments of ∼18–20 kDa are observed at all concentrations of PK, although smaller fragments appear with higher concentrations. No major differences in conformational subtype were demonstrated by this assay.
Association of PrP-129M with PrP-129V in Yeast
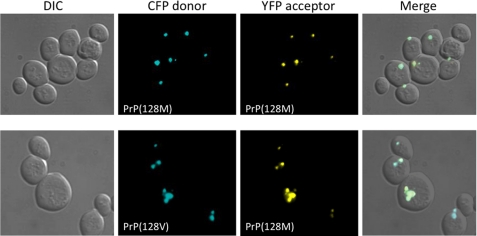
As a necessary prerequisite to the FRET studies, we expressed PrP-128M and PrP-128V as fluorescent fusion proteins, to confirm their potential to form aggregates and associate. Whereas expression of either CFP or YFP alone produced a diffusely uniform signal throughout the yeast cytosol, they produced distinct puncta, consistent with aggregate formation, when linked to the C-terminal of PrP23–230. Although a detailed analysis of the kinetics of aggregate formation was not undertaken, we observed no obvious qualitative differences between PrP-128M and PrP-128V aggregates (Fig. 3).
FIGURE 3.
Colocalization of PrP-128M and PrP-128V aggregates in the yeast cytosol. Differential interference contrast (DIC) (far left) and fluorescence images of yeast after 16 h of co-expression of PrP-128M and PrP-128V fluorophore pairs tagged with either CFP or YFP, as labeled. The far right column displays merged images. Both PrP fusion proteins produce dense aggregates. Co-expression of homologous or heterologous pairs produce >90% co-aggregation.
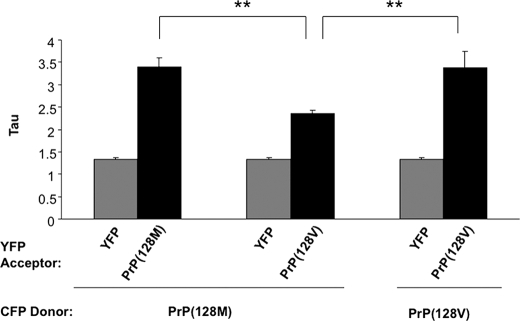
Next, homologous or heterologous PrP pairs were co-expressed and visualized by fluorescence microscopy. Co-expression of the homologous pair (PrP-128M::CFP and PrP-128M::YFP), hereafter designated PrP(128M/M) or the heterologous pair (PrP-128M::CFP and PrP-128V::YFP), designated PrP(128M/V), resulted in co-aggregation of greater than 90% of aggregates, suggesting that heterologous PrPs display similar aggregative behavior as homologous PrPs (Fig. 3). However, to determine whether the visualized PrP co-aggregates represent a composite of tightly associated heterologous PrP molecules or a coalescence of smaller aggregates composed of tightly associated homologous PrP, we applied live cell donor photobleaching FRET to this system. By measuring the decay of the donor fluorescence signal (PrP::CFP) after excitation, and fitting it with a single exponential decay curve, a Tau (τ) can be generated. A higher τ corresponds to a slower decay curve, indicating a stronger association between two interacting proteins, as time to quench the donor molecule is greater.
With each FRET pair analyzed, we determined the nonspecific background of a “non-FRET” pair, against which all FRET signals were compared. This was done by co-expressing CFP-tagged PrP-128M or PrP-128V with non-fused YFP, as YFP does not aggregate within the yeast cytosol nor is it predicted to specifically associate with PrP. Next, we measured FRET from yeast co-expressing PrP fused to YFP or CFP carrying the same (homologous pair) or different (heterologous pair) residues at 128. We found the τ for the homologous pair PrP(128M/M) was 3.39 ± 0.217, compared with 1.38 ± 0.062 for the non-FRET control pair PrP(128M/YFP), supporting a tight association of homologous PrP (Table 2, part A). In contrast, the τ for the heterologous PrP(128M/V) pair was 2.31 ± 0.159, lower (p < 0.01) than the homologous pair, yet higher (p < 0.001) than the negative control, suggesting a specific, but qualitatively different, association that formed between homologous PrPs (Table 2 and Fig. 4). Because the τ measured for the PrP(128V/V) pair was 3.37 ± 0.377, nearly identical to that measured for the PrP(128M/M) pair, the reduction in FRET observed with the PrP(128M/V) pair cannot be explained by a general alteration in the associative behavior of PrP-128V. These data, supported by the transmission data, clearly demonstrate an improved molecular association of misfolded PrP when residue 129 is homologous.
TABLE 2.
FRET of all PrP pairs
| Donor PrP (128X):CFP | Acceptor PrP(128X):YFP | Tau ± S.E. | Relative Tau ± S.E. |
|---|---|---|---|
| A. PrP(128M) and PrP(128V) | |||
| Met | 1.38 ± 0.062 | ||
| Met | Met | 3.39 ± 0.217 | |
| Met | Val | 2.31 ± 0.159 | |
| Val | Val | 3.37 ± 0.337 | |
| B. PrP(128X) non-FRET controls | |||
| Met | 1.35 ± 0.030 | ||
| Ala | 1.31 ± 0.063 | ||
| Leu | 1.42 ± 0.053 | ||
| Ile | 1.26 ± 0.041 | ||
| Tyr | 1.32 ± 0.077 | ||
| Val | 1.29 ± 0.047 | ||
| Mean = | 1.33 ± 0.023 | ||
| C. PrP(128X) and PrP(128X) | |||
| Met | Met | 3.98 ± 0.087 | 1.000 ± 0.022 |
| Ala | Ala | 3.94 ± 0.196 | 0.989 ± 0.049 |
| Leu | Leu | 3.79 ± 0.110 | 0.952 ± 0.027 |
| Ile | Ile | 4.58 ± 0.275 | 1.149 ± 0.069 |
| Tyr | Tyr | 4.43 ± 0.269 | 1.110 ± 0.068 |
| Val | Val | 3.68 ± 0.226 | 0.924 ± 0.023 |
| D. PrP(128X) and PrP(128V) | |||
| Val | Met | 2.35 ± 0.074 | 0.643 ± 0.020 |
| Val | Ala | 2.57 ± 0.251 | 0.702 ± 0.069 |
| Val | Leu | 2.59 ± 0.032 | 0.708 ± 0.009 |
| Val | Ile | 3.97 ± 0.243 | 1.09 ± 0.057 |
| Val | Tyr | 4.02 ± 0.266 | 1.03 ± 0.049 |
| Val | Val | 3.68 ± 0.226 | 1.00 ± 0.062 |
| E. PrP(128X) and PrP(128M) | |||
| Met | Met | 3.98 ± 0.087 | 1.000 ± 0.022 |
| Met | Ala | 3.76 ± 0.164 | 0.944 ± 0.041 |
| Met | Leu | 3.49 ± 0.101 | 0.877 ± 0.025 |
| Met | Ile | 2.53 ± 0.045 | 0.634 ± 0.011 |
| Met | Tyr | 2.19 ± 0.011 | 0.549 ± 0.010 |
| Met | Val | 2.35 ± 0.074 | 0.590 ± 0.019 |
FIGURE 4.
Live cell FRET analysis following co-expression of mouse PrP pairs homologous and heterologous at residue 128. PrP-128M::CFP was co-expressed with YFP, PrP-128M::YFP, or PrP-128V::YFP for 16 h and analyzed by FRET microscopy, as described under “Materials and Methods.” Actual Tau measures are plotted, and are recorded in Table 2. Higher Tau values represent higher FRET. Heterologous pairs displayed significantly reduced FRET compared with homologous pairs. Each bar represents the mean ± S.E. of a total of 35 to 50 separate measurements. **, p < 0.01.
Mutated Prion Propagation and Residue 129
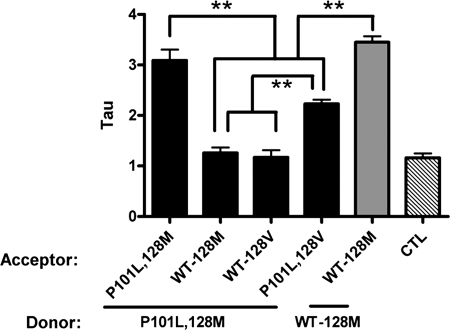
We next assessed the role of residue 129 in genetic prion disease. Prior transmission studies showed that Tg mice expressing a mouse-human chimeric PrP were resistant to the prion subtype of Gerstmann-Straussler-Scheinker disease (GSS), resulting from a P102L mutation of the PrP gene, whereas Tg mice that express low levels of PrP-P102L were susceptible to GSS(P102L), supporting homology at residue 102 as a necessary feature in the propagation of PrP-P102L prions (18). To model this intramolecular association, mouse PrP-P101L,128M was co-expressed with PrP-128M and analyzed. Strikingly, this pair displayed a τ of 1.26 ± 0.109, which was not different from the non-FRET control pair (τ = 1.16 ± 0.101), suggesting either absent, or a highly disrupted, association between PrP-P101L and WT-PrP (Fig. 5). However, when PrP-P101L,128M was co-expressed with the homologous PrP-P101L,128M partner, the association was strong (τ = 3.09 ± 0.248), paralleling the findings of the transmission studies. We then assessed the influence of residue 129 in the association of this mutant PrP. When PrP-P101L was co-expressed with PrP-P101L carrying a mismatch at residue 128, FRET was reduced (τ = 2.28 ± 0.085) to a level consistent with that measured for the WT heterologous PrP(128M/V) pair (τ = 2.31 ± 0.159).
FIGURE 5.
PrP-P101L associative behavior is dependent on residue 129. PrP-P101L,128M was co-expressed with PrP-P101L,128M, PrP-P101L, 128V, or WT-PrP with Met or Val at residue 128. PrP-P101L,128M was the donor (CFP tagged) and all others were acceptors (YFP tagged), as labeled. A control homologous pair of WT PrP-128M is represented by the gray bar. The CTL bar (diagonal lines) represents the τ value of the non-FRET control (PrP-P101L,128M paired with YFP), which was no different from the pairing with either WT-PrP. Each bar represents the mean ± S.E. of a total of 35 to 50 separate measurements. **, p < 0.001.
What Property of Residue 129 Determines PrP Association?
Based on the apparent predictive nature of this system, we employed it to address the question of whether the secondary structure propensities of Met and Val dictate the association of PrP molecules. To do this, we introduced a series of substitutions at residue 128 of mouse PrP and measured their associative behaviors. Although both Met and Val are hydrophobic non-polar amino acids, Met favors α-helical secondary structure and Val favors β-sheet, based on Chou-Fasman predictions (19). Thus, we substituted two non-polar amino acids that favor α-helical (Ala and Leu), and two that favor β-sheet (Ile and Tyr), secondary structure at position 128, one of which (Tyr) is slightly polar, and compared their associative behaviors with PrP-128M and PrP-128V.
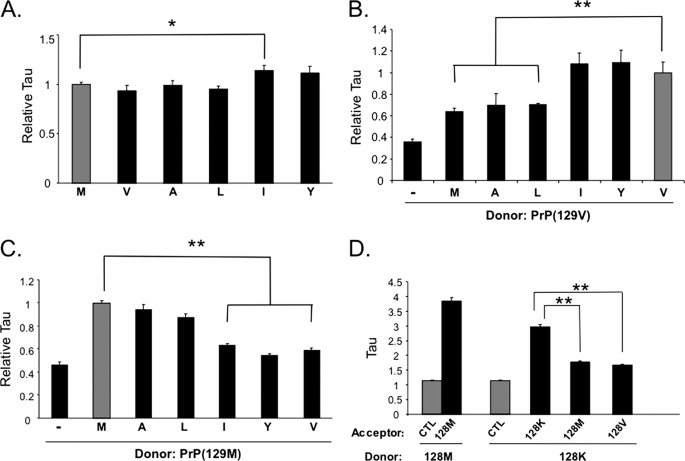
We first determined the τ for homologous PrP pairs for each 128 substitution to ensure they do not significantly alter the associative nature of PrP. A control τ was determined for each CFP-tagged PrP-128X, where X = Met, Val, Ala, Leu, Ile, Tyr, or Lys fused to CFP, paired with YFP alone (Table 2, part B). In practice, each of these control pairs was measured in parallel with one or more PrP pairs, to avoid any possible inherent variability between experimental runs. However, we found no statistical difference in the τ values among all control pairs (p > 0.05) and as such, the average of all control pairs (τ = 1.33 ± 0.023) was used as the baseline against which all τ values generated from PrP pairs were compared. Co-expression of each homologous pair resulted in highly reproducible and significant FRET, indicating these molecules undergo tight associations. The calculated τ values were: Ala = 3.94 ± 0.196, Leu = 3.79 ± 0.110, Ile = 4.58 ± 0.275, and Tyr = 4.43 ± 0.269 (Table 2, part C). The τ for each FRET pair was significantly higher than the background FRET measured either from the specific individual control pair PrP(128X/YFP) or the grouped mean of all control pairs. We normalized all pairs relative to the PrP(128M/M) pair and, interestingly, the Tyr and Ile substituted PrPs displayed relatively higher τ values than the homologous PrP(128M/M) pair, the latter attaining statistical significance, suggesting this β-inducing residue may associate more tightly than homologous PrP-128M molecules (p < 0.05) (Table 2, part C, and Fig. 6A).
FIGURE 6.
Comparative Tau values for all PrP(128X) paired with PrP-128M and PrP-128V. A, FRET of homologous PrP(128X/X) pairs. The Tau for each homologous PrP(128X/X) pair (black bars) are represented relative to WT PrP(128M/M) (gray bar). The label below each bar indicates the amino acid substitution at residue 128. PrP(128X)::CFP donor was co-expressed with its homologous PrP(128X)::YFP acceptor, and FRET performed on 35 to 50 separate co-aggregates for each substitution. Each homologous pair resulted in reproducible and significant FRET over a non-FRET control (see Table 1). B, relative Tau of heterologous PrP(128X/V) pairs (black bars) compared with the homologous PrP(128V/V) pair (gray bar). PrP-128V::CFP was the donor and PrP(128X)::YFP the acceptor, and X is each substitution, as labeled. The non-FRET PrP(128V/YFP) is also represented (−). C, relative Tau of heterologous PrP(128X/M) pairs (black bars) relative to the homologous PrP(128M/M) pair (gray bar), with PrP-128M::CFP as donor and PrP(128X)::YFP as acceptor, and X is each substitution, as labeled. The non-FRET PrP(128M/YFP) pair is also represented (−). D, Tau values for PrP-128K paired with PrP-128K, PrP-128M, or PrP-128V, compared with the homologous PrP(128M/M) pair (black bars) and the non-FRET control (gray bars), as labeled. For all experiments in this figure, *, p < 0.05; **, p < 0.001. All values, except those in panel D, are also provided in Table 1. M, Met; A, Ala; L, Leu; I, Ile; Y, Tyr; V, Val; K, Lys.
We next determined the τ values of each PrP-128X substitution paired with PrP-128M and PrP-128V. All τ values were significantly higher than the non-FRET control pair (p < 0.001), supporting a specific association with PrP-128V (Table 2, part D). When normalized relative to the PrP(128V/V) homologous pair, the τ of the PrP(128V/I) and PrP(128V/Y) pairs were not different, whereas those of the PrP(128V/A) and PrP(128V/L) pairs were significantly lower (p < 0.001) (Fig. 6B). In addition, the τ for each of the latter pairs were not significantly different from that of the PrP(128M/V) pair. Thus, PrP with Met, Ala, and Leu substitutions at 128 displayed similarly impaired associations with PrP-128V, whereas PrP with Ile and Tyr substitutions at 128 behaved the same as PrP-128V.
The above relationship held true when FRET was measured for the PrP(128M/X) pairs (Fig. 6C). In this case, the τ values for the PrP(128M/A) and PrP(128M/L) pairs were high, at 3.76 ± 0.164 and 3.49 ± 0.101, respectively, which compared well with the value of 3.98 ± 0.087 for the homologous PrP(128M/M) pair at this sitting (p > 0.05), whereas the τ values for the PrP(128M/I) and PrP(128M/Y) pairs were significantly lower (p < 0.001) (Table 2, part E). When normalized to the PrP(128M/M) pair, the τ values of the PrP(128M/A) and PrP(128M/L) pairs did not differ from each other or the PrP(128M/M) pair, whereas those measured for the PrP(128M/I) and PrP(128M/Y) pairs were significantly lower (Fig. 6C), but not different from PrP(128M/V), suggesting the latter pairs behaved in a similar manner as PrP(128M/V) heterologous pairs.
As a final test of this system, we assessed the importance of matching the non-polar and uncharged nature of Met and Val. We substituted the highly charged amino acid, Lys, and studied the associative behavior of it with wild-type PrP-128M and PrP-128V. The calculated τ value for the homologous PrP(128K/K) pair was 2.97 ± 0.079, supporting the potential of PrP(128K) molecules to specifically associate, although this value is significantly lower than that measured for the PrP(128M/M) pair at this sitting, supporting impaired association when this residue is charged. However, the τ for the PrP(128M/K) and PrP(128V/K) pairs were similar and significantly lower than that of the PrP(128K/K) pair (Fig. 6D), supporting a very altered or lack of association of these heterologous molecules, and confirming the importance of matching physical properties at position 129.
DISCUSSION
The precise mechanism by which PrPSc associates with PrPC to propagate PrPSc is only partially understood. Several lines of evidence support a more efficient PrPSc-PrPC association when the primary structures of these two species are identical at polymorphic residue 129 (5, 10, 15, 20). Although the structure of PrPSc is not known with certainty, it is believed that conversion from PrPC involves an α-helix to β-sheet transition at or near this site, and residue 129 may lie within the first new β-strand of PrPSc. Although epidemiologic data and prior transmission studies (5, 10) suggest that heterozygosity at codon 129 might confer protection from prion disease by impairing the physical association of PrPSc with PrPC, direct physical evidence for this has been lacking.
Our transmission results not only agree with the epidemiology of codon 129 in prion disease, they compliment the transmission results of others with vCJD, which are limited by the absence of the 129VV genotype (10), and confirm previous findings with sCJD (15). However, our FRET studies extend these results by providing compelling evidence that the physical association between PrPSc and PrPC molecules is likely to differ, depending on whether they are homologous or heterologous at residue 129. Our ability to detect measurable differences in FRET efficiency, rather than an “all or none” response, not only highlights the sensitivity of this model, but also suggests that the physical association between heterologous PrPs differ in a subtle, but specific way, from that of homologous PrPs. In addition, the reproducible nature of these data suggests the altered association of heterologous PrPs is an ordered, rather than random, process. Because FRET signal varies with 1/R6, where R is the distance between FRET partners, our data indicate that the C-terminal fluorophores are further apart when the FRET partners differ at residue 129. This impaired association of heterologous PrPs offers a plausible explanation, not only for the extended incubation periods observed when prions are transmitted across species, but also for the reported loss in fidelity of certain PrPSc conformations when PrPSc is transmitted to heterologous PrPC (10, 21).3
Thus, the identity of PrPSc and PrPC at residue 129 determines their interaction and efficiency of PrPSc propagation. In the yeast experiments, both species of PrP were expressed simultaneously and at similar levels. Microscopically apparent aggregates showed complete co-localization of the two proteins, even when these were non-identical at position 128 of mouse PrP. In this context, we also found that amino acids with similar physical properties and propensity for secondary structure as the native residue resulted in identical associative behaviors. Thus, Ile and Tyr, with predicted high β-sheet propensity (19), effectively substituted for Val, whereas Ala and Leu substitutions, predicted to promote α-helical structure, behaved identically to Met. These findings argue against a strict requirement for sequence homology, but instead suggest that the association of PrPSc with PrPC depends on their predicted propensities for a specific secondary structure. This hypothesis will best be tested by transmission studies in Tg mice carrying similar substitutions at residue 129.
Previous NMR studies of PrP91–231 have shown that PrP-128M and PrP-128V have very similar stability and structure (22), and this includes the side chains surrounding the polymorphic site. Nevertheless, the structure of PrPSc differs from that of PrPC, which these authors studied. The sites at which PrPC binds to PrPSc, and through which conversion to PrPSc occurs, are likely to be dynamic, and because NMR is an ensemble technique, rare structures may not be observed. It is not known how this transition occurs, but modeling and transmission studies suggest that it involves a template-induced structural transition (13, 23). It is possible that within the dynamic ensemble of structures surrounding residue 129, those with β-sheet structure at this site in PrPC have enhanced affinity for a homologous site on the surface of PrPSc. This is supported by the enhanced susceptibility of Tg(129V) mice, when challenged with PrPSc-129M, compared with the significantly reduced susceptibility of Tg(129M) mice challenged with PrPSc-129V. It is of interest that the yeast studies revealed higher FRET values of homologous PrP-128Y and PrP-128I compared with PrP-128M, and even PrP-128V, suggesting that those residues might increase the propensity of this site to form β-sheet. Overall, the FRET studies, in concert with the transmission results reported here and elsewhere (10, 15) support a model in which the association of PrPSc with PrPC depends intimately on the pairing of residue 129 in each, to result in a high fidelity and efficiency of conformational transfer.
The utility of this model to predict the association of heterologous PrP molecules is underscored by the studies with PrP-P101L, in which FRET was absent between PrP-P101L-128M and WT PrP-128M, but high between homologous PrP101L-128M molecules. These results correlate directly with transmission studies that found Tg mice that express WT-PrP were resistant to GSS(P102L), whereas those expressing GSS(P102L) at low levels were susceptible (18). Our findings suggest the interaction of PrPSc with PrPC appears to be more dependent on homology at residue 102 than residue 129, because co-expression of PrP carrying the mouse P101L mutation with heterologous sequences at residue 128 displayed intermediate FRET. Thus, a mismatch at residue 102 results in a complete barrier to transmission because of the absence of any degree of an ordered association, whereas a mismatch at residue 129 produces a partial barrier that results from an altered association. These results agree with early reports that PrP amyloid deposits in GSS(P102L) are comprised of only mutant PrP and not WT-PrP (24). They also suggest that, in contrast to sCJD, GSS(P102L) is not affected by the status of codon 129 on the WT allele.
In summary, our live cell FRET model to study the associative properties of heterologous PrPs predicts that the physical association of PrPSc with PrPC is directly affected by a mismatch in the primary structure of each, and the degree to which the interaction is altered depends on the specific residue involved, primarily with respect to its predicted secondary structure. The remarkable correlation of these conveniently obtained in vitro findings with the lengthy and costly rodent transmission studies, suggests a novel alternative with which to assess the transmissible potential of prions within and among different species and humans, and to aid in defining the interface between PrPSc and PrPC.
Acknowledgments
We thank Stanley Prusiner, M.D. (University of California, San Francisco) for the kind gift of Tg440 and Tg152 breeders that were used to generate our mouse lines and Julie Cook for technical work.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS046037 and R01NS051480, The Brain Research Foundation, The Pioneer Fund, and a gift from Barbara and Marc Posner.
S. Mallik and J. A. Mastrianni, unpublished data.
- PK
- proteinase K
- FRET
- fluorescence resonance energy transfer
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- GSS
- Gerstmann-Straussler-Scheinker
- CJD
- Creutzfeldt-Jakob disease
- Tg
- transgenic
- Hu
- human
- WT
- wild-type.
REFERENCES
- 1.Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mastrianni J. A., Roos R. P. (2000) Semin. Neurol. 20, 337–352 [DOI] [PubMed] [Google Scholar]
- 3.Moore R. A., Vorberg I., Priola S. A. (2005) Arch. Virol. Suppl. 19, 187–202 [DOI] [PubMed] [Google Scholar]
- 4.Telling G. C., Scott M., Hsiao K. K., Foster D., Yang S. L., Torchia M., Sidle K. C., Collinge J., DeArmond S. J., Prusiner S. B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9936–9940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owen F., Poulter M., Collinge J., Crow T. J. (1990) Am. J. Hum. Genet. 46, 1215–1216 [PMC free article] [PubMed] [Google Scholar]
- 6.Parchi P., Capellari S., Chin S., Schwarz H. B., Schecter N. P., Butts J. D., Hudkins P., Burns D. K., Powers J. M., Gambetti P. (1999) Neurology 52, 1757–1763 [DOI] [PubMed] [Google Scholar]
- 7.Collinge J., Palmer M. S., Campbell T., Sidle K. C., Carroll D., Harding A. (1993) Br. Med. J. 306, 301–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mead S. (2006) Eur. J. Hum. Genet. 14, 273–281 [DOI] [PubMed] [Google Scholar]
- 9.Mead S., Poulter M., Uphill J., Beck J., Whitfield J., Webb T. E., Campbell T., Adamson G., Deriziotis P., Tabrizi S. J., Hummerich H., Verzilli C., Alpers M. P., Whittaker J. C., Collinge J. (2009) Lancet Neurol. 8, 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wadsworth J. D., Asante E. A., Desbruslais M., Linehan J. M., Joiner S., Gowland I., Welch J., Stone L., Lloyd S. E., Hill A. F., Brandner S., Collinge J. (2004) Science 306, 1793–1796 [DOI] [PubMed] [Google Scholar]
- 11.Bishop M. T., Hart P., Aitchison L., Baybutt H. N., Plinston C., Thomson V., Tuzi N. L., Head M. W., Ironside J. W., Will R. G., Manson J. C. (2006) Lancet Neurol. 5, 393–398 [DOI] [PubMed] [Google Scholar]
- 12.Norstrom E. M., Mastrianni J. A. (2005) J. Biol. Chem. 280, 27236–27243 [DOI] [PubMed] [Google Scholar]
- 13.Mastrianni J. A., Nixon R., Layzer R., Telling G. C., Han D., DeArmond S. J., Prusiner S. B. (1999) N. Engl. J. Med. 340, 1630–1638 [DOI] [PubMed] [Google Scholar]
- 14.Li X., Rowland L. P., Mitsumoto H., Przedborski S., Bird T. D., Schellenberg G. D., Peskind E., Johnson N., Siddique T., Mesulam M. M., Weintraub S., Mastrianni J. A. (2005) Ann. Neurol. 58, 858–864 [DOI] [PubMed] [Google Scholar]
- 15.Korth C., Kaneko K., Groth D., Heye N., Telling G., Mastrianni J., Parchi P., Gambetti P., Will R., Ironside J., Heinrich C., Tremblay P., DeArmond S. J., Prusiner S. B. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4784–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kascsak R. J., Tonna-DeMasi M., Fersko R., Rubenstein R., Carp R. I., Powers J. M. (1993) Dev. Biol. Stand. 80, 141–151 [PubMed] [Google Scholar]
- 17.Ma J., Lindquist S. (1999) Nat. Cell Biol. 1, 358–361 [DOI] [PubMed] [Google Scholar]
- 18.Telling G. C., Haga T., Torchia M., Tremblay P., DeArmond S. J., Prusiner S. B. (1996) Genes Dev. 10, 1736–1750 [DOI] [PubMed] [Google Scholar]
- 19.Chou P. Y., Fasman G. D. (1974) Biochemistry 13, 222–245 [DOI] [PubMed] [Google Scholar]
- 20.Parchi P., Giese A., Capellari S., Brown P., Schulz-Schaeffer W., Windl O., Zerr I., Budka H., Kopp N., Piccardo P., Poser S., Rojiani A., Streichemberger N., Julien J., Vital C., Ghetti B., Gambetti P., Kretzschmar H. (1999) Ann. Neurol. 46, 224–233 [PubMed] [Google Scholar]
- 21.Kobayashi A., Asano M., Mohri S., Kitamoto T. (2007) J. Biol. Chem. 282, 30022–30028 [DOI] [PubMed] [Google Scholar]
- 22.Hosszu L. L., Jackson G. S., Trevitt C. R., Jones S., Batchelor M., Bhelt D., Prodromidou K., Clarke A. R., Waltho J. P., Collinge J. (2004) J. Biol. Chem. 279, 28515–28521 [DOI] [PubMed] [Google Scholar]
- 23.Telling G. C., Parchi P., DeArmond S. J., Cortelli P., Montagna P., Gabizon R., Mastrianni J., Lugaresi E., Gambetti P., Prusiner S. B. (1996) Science 274, 2079–2082 [DOI] [PubMed] [Google Scholar]
- 24.Tagliavini F., Lievens P. M., Tranchant C., Warter J. M., Mohr M., Giaccone G., Perini F., Rossi G., Salmona M., Piccardo P., Ghetti B., Beavis R. C., Bugiani O., Frangione B., Prelli F. (2001) J. Biol. Chem. 276, 6009–6015 [DOI] [PubMed] [Google Scholar]