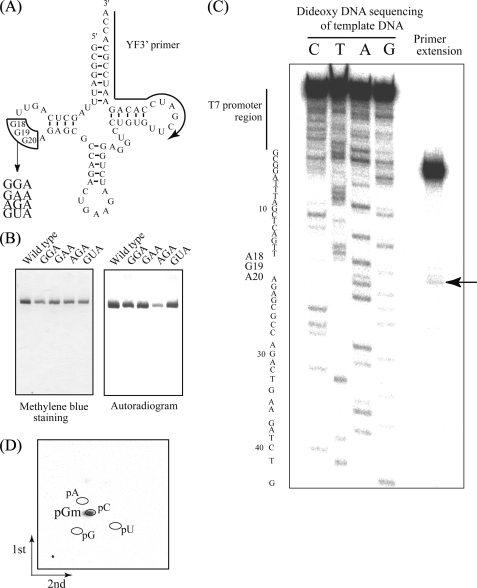
FIGURE 3.
A, substitution sites (G18G19G20) and replaced sequences depicted on a tRNAPhe clover-leaf schematic. The substitution site (G18G19G20) is enclosed by lines, and the region complementary to the primer used for extension (C) is marked with an arrow. B, methylation of tRNAPhe variants. Methyl group acceptance activities of tRNAPhe variants GGA, GAA, AGA, and GUA were analyzed using the same method as Fig. 2C. C, primer extension to locate sites of modification. Following methylation of tRNAPhe AGA using TrmH and AdoMet, primer extension was performed as described under “Experimental Procedures.” To generate the DNA sequence ladder on the left, dideoxy DNA sequencing was performed using the same 5′-32P-labeled primer. The terminated band (marked with an arrow) shows 2′-O-methylation at G19. D, two-dimensional thin layer chromatography analysis of the modified nucleotide. The tRNA AGA variant was methylated using TrmH and [14C]AdoMet, digested with nuclease P1, and then separated on two-dimensional thin layer chromatography plates prior to autoradiography.