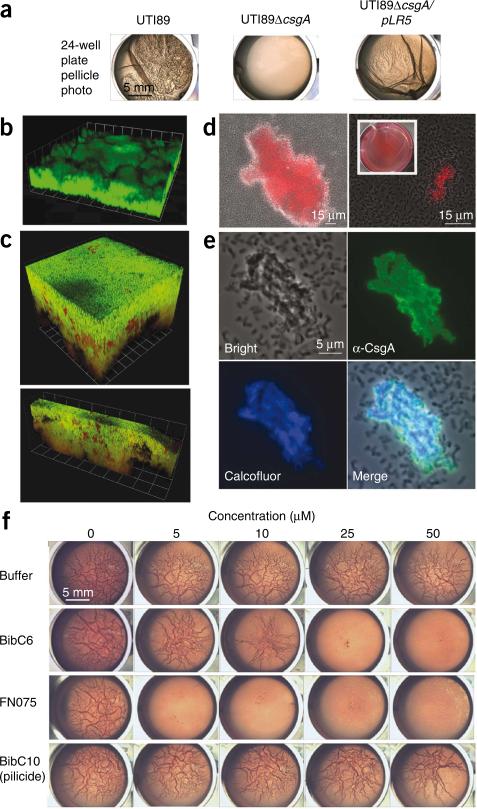
Figure 3.
Curlicide inhibition of curli-dependent UTI89 pellicle formation. (a) Macroscopic observation of curli-dependent pellicles. Pellicle biofilms were visible in wild-type UTI89 culture 48 h post-inoculation in YESCA broth. Deletion of csgA (UTI89ΔcsgA) abolished pellicle formation, and the pellicle was restored by providing csgA in trans (UTI89ΔcsgA/pLR5). (b) Three-dimensional confocal microscopy reconstructions revealed thick pellicles with complex architectures. Pellicles were stained with nucleic acid dye STYO9 (green). Each grid unit is 14.3 μm. (c) Three-dimensional reconstructions of confocal microscopy images showed that GFP reporter gene expression (green) driven by the csgB; csgA promoter is active in UTI89 pellicle. Bacteria were counterstained with nucleic acid dye SYTO83 (red). Grid units were 7.4 μm for the top panel and 14.3 μm for the bottom panel. (d) Curli were detected in the pellicle by Congo red uptake in bright field (DIC) images merged with the red fluorescence channel. Congo red uptake was apparent by macroscopic examination of pellicle (inset). (e) Immunostaining of pellicle segments with anti-CsgA antisera illustrated homogeneous CsgA presence among aggregated cells, whereas planktonic bacteria were unlabeled (top right panel). Calcofluor staining indicated that β-glucans such as cellulose colocalize in the pellicle (bottom panels). (f) Curlicides blocked pellicle formation in a 24-well plate assay in which UTI89 was grown in Congo red–containing YESCA medium at 30 °C (rows 2 and 3). Congo red had no effect on the results but permitted facile display of pellicle roughness. Equivalent carrier volumes of ethanol had no effect on pellicle formation (top row), and pellicle was unaffected by pilicide BibC10, 1 (bottom row).