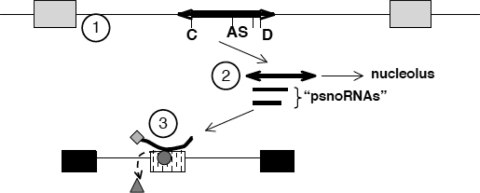
Figure 6.
Model for MBII-52 action on RNA processing. (1) The PWS critical region contains snoRNAs (thick line) located in introns between non-coding exons (grey boxes). The snoRNA is characterized by a C box (C), D box (D) and an antisense box (AS), as well as stem-forming sequences (arrows). (2) This unit generates several RNAs, including the full-length snoRNA that shows its highest concentration in the nucleolus and Cajal bodies (35) as well as several shorter psnoRNAs (for processed snoRNAs). PsnoRNAs are present in the nucleoplasm where they associate with hnRNPs. (3) PsnoRNAs can change splice-site selection, most likely by binding to complementary sequences. We postulate that they either remove regulatory proteins from their targets (triangle) or bring in associated proteins to the exon recognition complex (diamond associated with the small RNA).