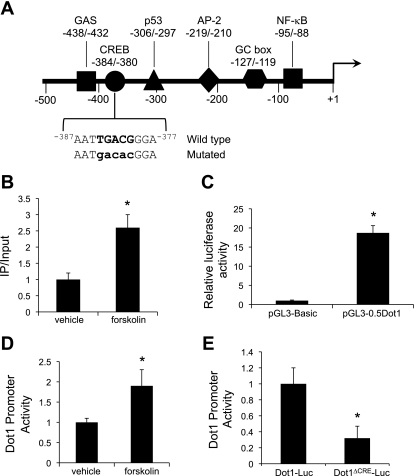
Fig. 3.
A: map of the proximal 5′-flanking region of the murine Dot1 gene. Consensus sites for the binding of selected transcription factors are indicated. Sequence of the −384/−380 region and the mutated sequence (mutations in lower case letters) used for trans-activation assays are also indicated. B: ChIP-qPCR analysis of CRE-binding protein (CREB) occupancy at the Dot1 proximal promoter. MMCs were treated with vehicle or forskolin (1 μM) for 24 h. ChIP assays were then performed with antibody directed against CREB or nonimmune IgG followed by qPCR with a primer set designed to amplify −582 to −162 of the Dot1 gene. The value for the final amplicon/input DNA with anti-CREB from the vehicle-treated cells was set as 1, and the value from the forskolin-treated cells was normalized to it. Values are means ± SE of 3 separate experiments, each performed in triplicate. C: luciferase assay showing functional promoter activity of the transiently transfected pGL3-0.5Dot1 construct. pGL3-Basic or pGL3-0.5Dot1 was cotransfected with pRL-SV40 into MMCs, and firefly and Renilla luciferase activities were measured in triplicate for each sample. Firefly luciferase activity was normalized to that of Renilla luciferase activity to generate “Dot1 promoter activity.” *P < 0.05 vs. pGL3-Basic-transfected cells, n = 3. D: luciferase activity showing forskolin (1 μM for 24 h) induction of pGL3-0.5Dot1. MMCs were cotransfected with pGL3-0.5Dot1 and pRL-SV40 in MMCs, and the cells were treated with vehicle or forskolin (1 μM) for 24 h. Firefly luciferase activity was normalized to that of Renilla luciferase activity to generate “Dot1 promoter activity,” measured in triplicate for each sample. *P < 0.05 vs. vehicle-treated cells, n = 3. E: functional analysis of the −384/−380 CRE in the proximal promoter of the murine Dot1 gene. Core nucleotides of the CRE in the pGL3-0.5Dot1 were mutated by point mutagenesis to create pGL3-0.5Dot1Δ−384/−380 (Dot1Δ−384/−380-Luc) as described in materials and methods. The construct or pGL3-0.5Dot1 was cotransfected with pRL-SV40 into MMCs, and firefly and Renilla luciferase activities were measured in triplicate for each sample. Firefly luciferase activity was normalized to that of Renilla luciferase activity to generate Dot1 promoter activity. Error bars indicate ±SE. *P ≤ 0.05 vs. pGL3-0.5Dot1 (Dot1-Luc), n = 3.